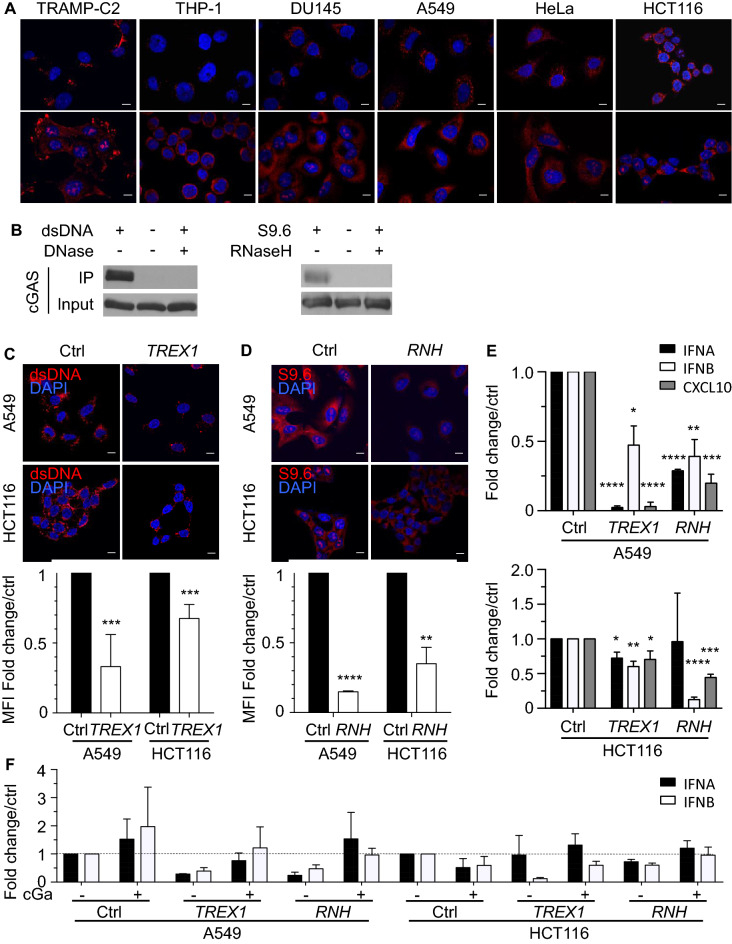
Figure 2.
Cytosolic DNA Levels Do Not Contribute to the STING Dysfunction in Human Cancer Cells. (A) TRAMP-C2, THP-1, DU145, A549, HeLa, and HCT116 cells were stained for dsDNA or RNA:DNA hybrids recognized by the S9.6 antibody (red) in presence of DAPI (blue). (B) Cytosolic dsDNA (left panel) and RNA:DNA hybrids (right panel) in A549 cell lysates were immunoprecipitated with dsDNA-specific or S9.6 antibodies. Precipitates were immunoblotted using a cGAS-specific antibody. A portion of the lysates was treated with 20 units DNase or RNaseH prior to elution. The shown blots are representative of 3 independent experiments. (C) A549 or HCT116 cells were transduced with a retrovirus encoding TREX-1 or control vector and were stained for dsDNA (red) in presence of DAPI (blue). Bar graph shows quantification of the dsDNA MFI ± SD in TREX1- (white column) and control- (black column) transduced A549 and HCT116 cells. Values were normalized to the MFI in control cells. See also Figure S3. (D) RNASEH1 (RNH)- or control-transduced A549 and HCT116 cells were stained for RNA:DNA hybrids (red) in the presence of DAPI (blue). Bar graph shows quantification of the RNA:DNA hybrid MFI ± SD in RNASEH1 (RNH)- (white column) and control- (black column) transduced A549 and HCT116 cells. Values were normalized to the MFI in control cells. See also Figure S3. (E) IFNA (black column), IFNB (white column) and CXCL10 (grey column) transcript levels in TREX1- or RNASEH1 (RNH)-transduced A549 and HCT116 cells were determined by real-time PCR. Transcript levels were normalized to transcript levels in control-transduced cells. (F) TREX1- and RNASEH1 (RNH)-transduced A549 and HCT116 cells were treated with PBS (Ctrl) or 2 μg/ml cGAMP (cGa) for 4 h. IFNA (black column) and IFNB (white column) transcript levels were measured by real-time PCR and normalized to control cells. Scale bar denotes 10 μm in all images. All data are presented as mean ± SD of 3 independent experiments.