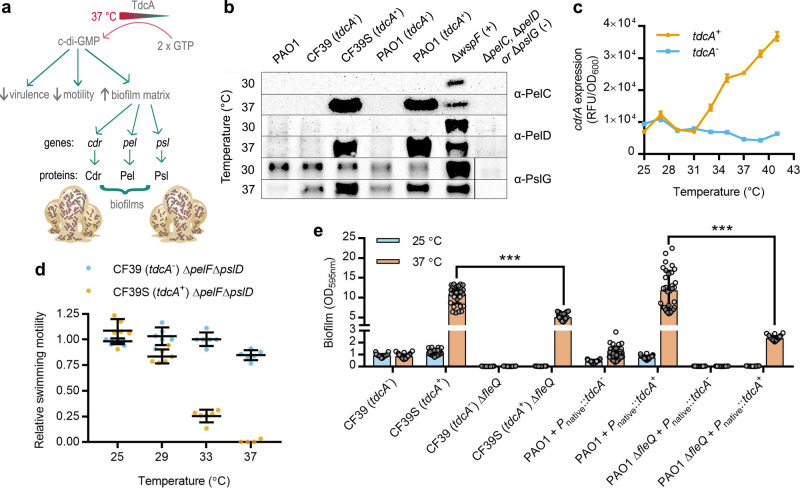
Fig. 2. Cyclic diguanylate (c-di-GMP)-dependent thermotransduction.
a Schematic of the heat-activated c-di-GMP pathway that drives biofilm development in P. aeruginosa CF39S. b Western blots for c-di-GMP-regulated Pel and Psl proteins. Measurements were performed in biological triplicate and one representative blot is shown (original, unedited blot images are provided in Supplementary Information). Lanes were loaded with a normalized quantity of total cellular protein. The ΔwspF strain, which has constitutively high c-di-GMP levels23, 32, was used as a positive control. c Expression of the PcdrA::gfp reporter. Data represent means and standard deviations of measurements each performed in biological and technical triplicate. The tdcA− and tdcA+ strains are represented by sky blue and orange lines, respectively. OD600nm denotes the optical density measured at a wavelength of 600 nm, and RFU denotes relative fluorescence units. d Thermal control of swimming motility in P. aeruginosa CF39S. Measurements were made in ΔpelF ΔpslD backgrounds to limit interference from extracellular polymers in motility phenotypes. Each datum point represents an independent biological replicate, and lines and bars represent means and standard deviations, respectively, for 4–6 independent biological replicates; tdcA− and tdcA+ strains are represented by sky blue and orange dots, respectively. e Biofilm assays indicate that TdcA integrates into the P. aeruginosa c-di-GMP regulatory network, in part, via the c-di-GMP-binding transcription factor, FleQ. Datum points represent 12 technical replicates from each of three biological replicates, and lines and bars represent means and standard deviations, respectively. Temperatures of 25 °C and 37 °C are represented by sky blue and orange bars, respectively. OD595nm denotes the optical density measured at a wavelength of 595 nm. Significant difference via two-tailed Student’s t-test (*** denotes P < 0.001). Strains denoted tdcA− have the tdcA162ΔG allele.