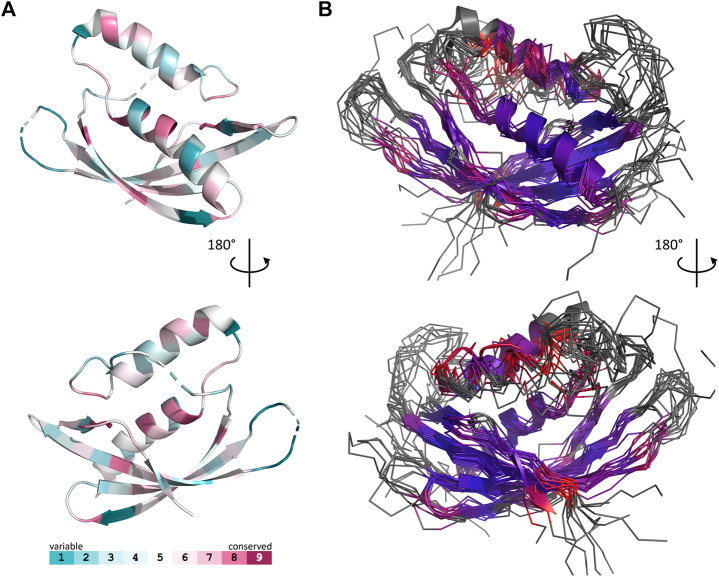
FIGURE 3.
Comparison of 17 representative three-dimensional structures of VirB8 and VirB8-like proteins. The structure of OrfG64–204 (PDB id 6zgn) is represented as a cartoon (A), with its amino acids colored according to the Consurf server (Ashkenazy et al., 2016) which analyzed the similarity calculated in the sequence alignment deduced from the structure superposition produced by mTM-Align (Dong et al., 2018a) for a set of sventeen selected proteins. The amino acid shown with sticks (Val191 in OrfG) and localised in β4 facing α 1 is the only one whose side chain shares a similar nature in all structures, while there is no conserved residue. The 17 structures (PDB ids 2cc3, 3ub1 (limited to TcpC104–231), 3wz3, 3wz4, 4akz, 4ec6, 4jf8, 4kz1, 4lso, 4mei, 4nhf, 4o3v, 5aiw, 5cnl, 5i97, 6iqt, 6zgn) are represented (B) as superposed wires (except OrfG as a cartoon) using a color-code based on the quality of the superposition of these structures. For each position, a gradient from violet to red is used depending on the low to high values of the root mean square calculated on the distance between all the possible pairs of the 17 superposed Cα carbons at this position (grey means that mTM-Align found at least one structure impossible to align). This representation highlights the resemblances and differences in the NTF2-like folds of these proteins.