Figure 3.
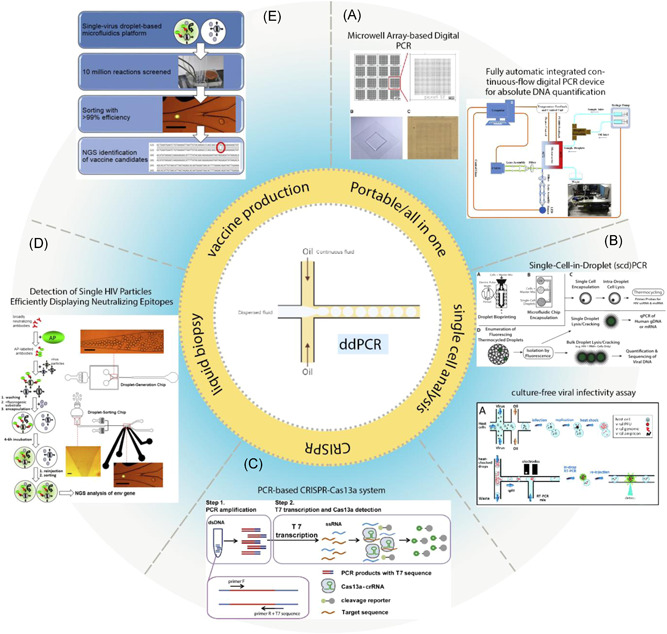
Integration of droplet‐based microfluidic in various biological methods. (A) This figure shows a microwell array‐based digital PCR made by conventional soft lithography, Illustrated by a fluorescent microscope. The use of the microwell Array reduces generation, thermal cycling, and reading of droplets cost. 169 Schematic diagram of a fully automatic integrated digital PCR device based on continuous‐flow digital PCR works with solar power. (B) Schema of the single‐cell encapsulation, lysis, Tat‐Rev spliced or unspliced cell‐associated HIV‐1 RNA detection, and rescue of cellular genomic DNA and mRNA in droplets. Droplets are created by one of the bio print methods or microfluidic chips. CD4+ T‐cells were isolated from peripheral blood mononuclear cells (PBMCs). CD4+ T cells were washed and mixed 1:1 with ddPCR master mix and cell lysis agent. Droplets generated, and cells are lysed inside the droplets, followed by PCR amplification of Tat‐Rev spliced or unspliced cell‐associated HIV‐1 RNA. 170 The schematic diagram shows a rapid, targeted, and culture‐free viral infectivity assay in drop‐based microfluidics. murine norovirus strain MNV‐1 that infect host cells, coencapsulated in droplets. 147 (C) PCR‐based CRISPR‐Cas13a system. 171 (D, E) A highly efficient microfluidic droplet‐based platform to screen single virus particles for optimum vaccine candidate antigenic compounds. Like the standard ELISA method, HIV‐1 particles were incubated with alkaline phosphatase (AP)‐labeled broadly neutralizing antibody PGT128. Next, HIV‐1 particles were bonded to AP were encapsulated separately in droplets, together with a fluorogenic substrate for AP (FDP). Finally, the fluorescent intensity of each droplet was measured and sorted by another microfluidic device. Viruses were recovered, and NGS of the env genes was performed after RT‐PCR using specific primers for HIV‐1 env. 175 172 ELIS, enzyme‐linked immunosorbent assay; mRNA, messenger RNA
