Figure 1.
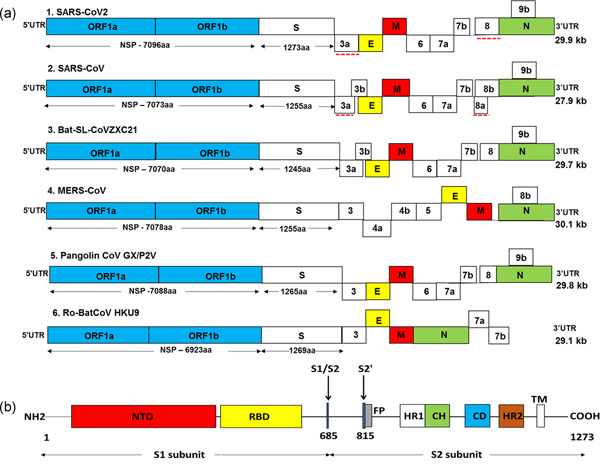
(a) Genome structure of SARS‐CoV‐2 and other coronaviruses. The genome of CoVs comprises of 5ʹ and 3ʹ untranslated region (UTR) and open reading frame (ORF) 1a/b (blue boxes). The structural genes present at 3ʹ terminus encodes for the structural proteins including spike (S; white boxes), envelope (E; yellow boxes), membrane (M; red boxes), and nucleocapsid (N; green boxes) which are common features to all CoVs. In addition, the accessory genes interspaced between the structural genes encodes for accessory proteins. The comparison of coding regions of SARS‐CoV‐2 with different CoVs showed a similar genome organization to SARS‐CoV, bat SL‐CoVZXC21, and pangolin CoV GX/P2V. There is no remarkable difference in the ORF1 of different CoVs but it encodes for NSPs of variable lengths and there is a distinction in the accessory genes. The red dotted line shows a notable variation between SARS‐CoV‐2 and SARS‐CoV. Red dotted line: notable variation between SARS‐CoV‐2 and SARS‐CoV. (b) SARS‐CoV‐2 spike (S) glycoprotein. The S1 region of spike protein contains a N‐terminal domain (NTD; red box) and a C‐domain or receptor‐binding domain (RBD; yellow box). The S2 subunit contains the fusion peptide (FP; gray box), heptad repeat 1 (HR1; white box), central helix (CH; green), connector domain (CD; blue box), heptad repeat 2 (HR2; brown box), and transmembrane domain (TM). Black arrows: cleavage sites at S1/S2 boundary (R685) and S2ʹ (R815)
