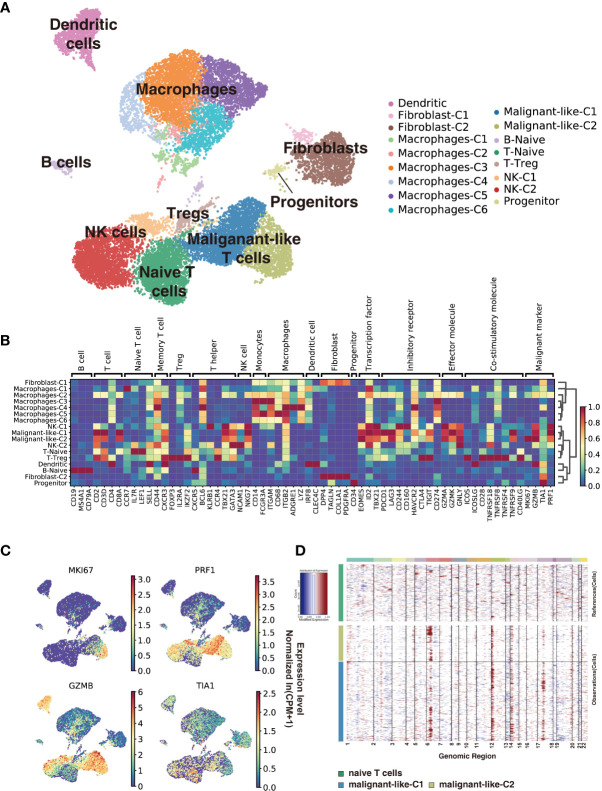
Figure 2.
Subcutaneous panniculitis-like T-cell lymphoma ecosystem at single-cell resolution. Cells from the patient’s subcutaneous lesion tissue were clustered using the Leiden community detection algorithm to identify groups of cells with similar expression patterns. (A) Single-cell expression of the subcutaneous lesions’ cells in UMAP space (first two dimensions). Cells are color-coded according to the clusters generated by the Leiden algorithm. (B) Heatmap summarizes the mean expression (normalized and log-transformed) of selected canonical markers in each cluster. The gene expression value has been scaled for visualization. The covariate bar on the top side indicates the component associated with each gene, and red boxes highlight the prominent expression of genes for the known subtypes. (C) UMAP plots of malignant markers (MKI67, PRF1, TIA1, GZMB) expression in subcutaneous lesions’ cells. (D) Chromosomal landscape of inferred large-scale copy number variations (CNVs) distinguishes malignant from non-malignant cells. Amplifications (red) or deletions (blue) were inferred by averaging expression over 100-gene stretches on the respective chromosomes.