Figure 2. Gly‐ and diGly‐ending proteins are scarce in functional proteomes.
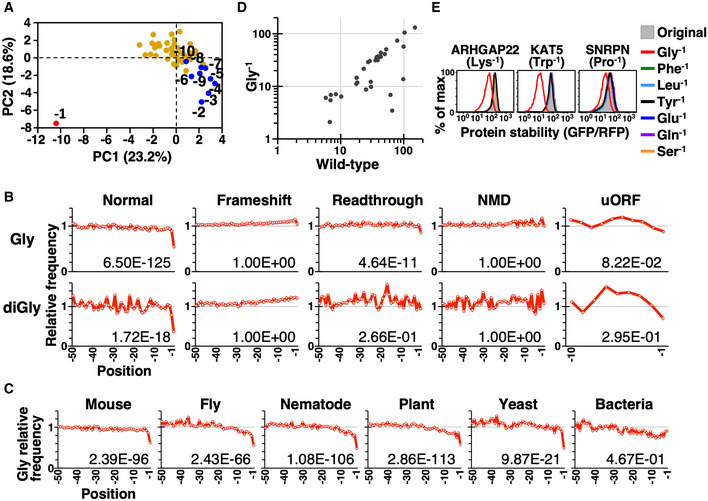
- PCA on the amino acid frequency of the last 50 residues of functional human proteins. Human protein coding sequences were downloaded from NCBI GeneBank. The terminal (−1) and the final 10 residues (−2 to −10) are labeled red and blue, respectively, with the remainder marked yellow.
- The relative frequency of Gly (top) and diGly (bottom) in the final 50 residues of functional and aberrant human proteins. We only analyzed the last 10 residues of peptides translated from uORFs due to their inherently smaller size. The number on each graph indicates the left‐tailed FDR (false discovery rate)‐adjusted P‐value of the extreme terminal (−1) residue. Analyses using human sequences downloaded from Uniprot database are shown in Fig EV1C and D.
- The relative frequency of Gly at the C‐termini of indicated proteomes, i.e., mouse (Mus musculus), fly (Drosophila melanogaster), nematode (Caenorhabditis elegans), plant (Arabidopsis thaliana), yeast (Saccharomyces cerevisiae), and bacteria (Escherichia coli).
- Stability (mean GFP/RFP) comparisons between the wild‐type protein and its mutant in which the terminal residue has been changed to Gly. Detailed information on tested proteins is presented in Table EV3.
- Comparisons of protein stability among wild‐type protein and the mutants indicated on the right.
