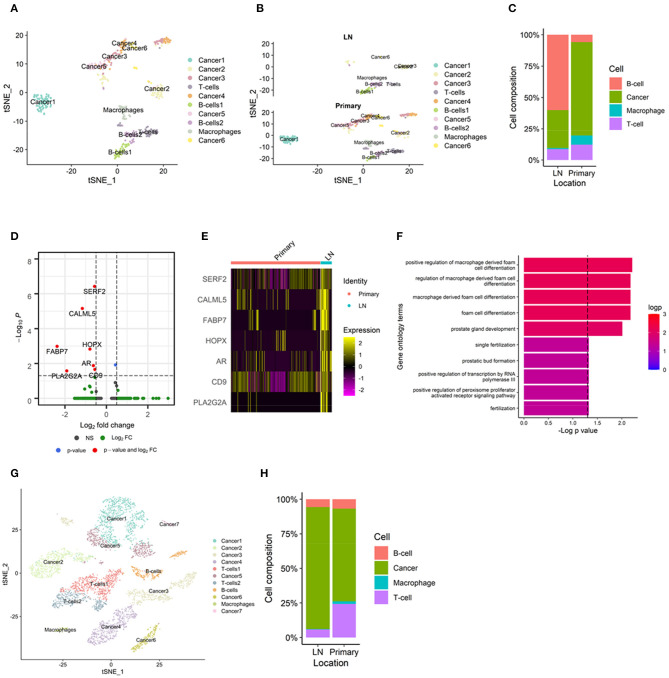
Figure 4.
TIME of metastatic lymph nodes analyzed by single-cell RNA-sequencing. (A) A total of 515 cells from an scRNA-seq dataset clustered into 10 clusters and 4 cell types. (B) There were fewer macrophages in metastatic lymph nodes than in primary tumors in the t-SNE plot. (C) Bar plot showing macrophages in metastatic lymph nodes and primary tumors. (D) Volcano plots representing seven differentially expressed genes: SERF2, CALML5, FABP7, HOPX, AR, CD9, and PLA2G2A. All the seven DEGs showed negative log2 fold change; lower expression in primary tumors than in metastatic lymph nodes. (E) Heatmap demonstrating the expression of each differentially expressed gene. (F) In gene ontology analyses, macrophage-related terms were selected as significant. (G) In a head and neck cancer scRNA-seq dataset, there were fewer macrophages in metastatic lymph nodes than in primary tumors in the t-SNE plot. (H) Bar plot showing macrophages in metastatic lymph nodes and primary tumors. AR, androgen receptor; CALML5, calmodulin-like protein 5; FABP7, fatty acid binding protein 7; HOPX, homeodomain-only protein; PLA2G2A, phospholipase A2 group IIA; scRNA-seq, single-cell RNA sequencing; SERF2, small EDRK-rich factor 2; TIME, tumor immune microenvironment; t-SNE, t-distributed stochastic neighborhood embedding.