Despite news about the new virus SARS‐CoV‐2 and COVID‐19 grabbing the headlines in the beginning of 2020, life and work carried on as normal in most academic labs. Lab projects were the focus, results discussed face‐to‐face and hypothesis dismissed. Conferences were attended in person and flights to the next meeting often booked already. By March, the seriousness became clear. With surrounding countries reporting increasing number of infections, the emergence of SARS‐CoV‐2 in Portugal was a matter of time, with the first two Portuguese cases reported at the start of March. Newspapers reported the cases with warnings not to be alarmist and that officials would calmly follow the evolution of the events (see here). However, how do you follow the spread of a virus, especially when it is new and capacity to detect it is limited?
Just a little help
By the middle of March, the first fatal COVID‐19 case was reported at Hospital Santa Maria (HSM) in Lisbon (see here). Health authorities reported limited test capacities nationwide, and local entities such as hospitals did not have the capacity to test the patients and staff required. The Instituto de Medicina Molecular João Lobo Antunes (iMM, https://imm.medicina.ulisboa.pt/), is a basic and translational life sciences research institute located in Lisbon on the site of the largest hospital in Portugal, the HSM, home also of the Medical School of Lisbon (Faculdade de Medicina da Universidade de Lisboa). The interactions with the hospital and medical school are close and several clinicians hold research positions at iMM. Clinical colleagues reported the high demand and the limited options available to detect this new virus.
Detecting a virus is not hard for someone working in the life sciences. A virus has unique genome material: “It is just a polymerase chain reaction or PCR!” some of us in the lab thought. A small team of iMM researchers met with doctors at HSM in the second week of March to discuss how we could assist with diagnostics. Of course, viral detection is a bit more complex than just a PCR. SARS‐CoV‐2 is a RNA virus, requiring the need for reverse transcription to generate, the more stable and easier to amplify, DNA. The bottleneck was the isolation of viral RNA from samples. On 11th March 2020, the World Health Organisation (WHO) has declared COVID‐19 as a pandemic (see here). The handling of potential contaminated material required laboratory safety level (BSL)‐3 making testing extra difficult. It soon transpired that with increasing sample numbers requiring rapid analysis, more help was needed.
National and local measures to control viral spread were introduced rapidly and involved the closure of the iMM building on March 13th. Seminars and lab meetings were cancelled, any travel was strongly discouraged, and a video call platform entered the collective psyche for the first time. Effectively, a large amount of hardworking and talented scientists found themselves contemplating which series to watch on video‐streaming services, which books to read, and an occasional daring one started to prepare for that DIY job postponed so often already. However, the iMM quarantine did not last long; about 120 people enthusiastically met a call for volunteers to set up COVID‐19 diagnostics the following week. The desire to help the country overcome the pandemic prompted the iMM community, from all career levels, to repurpose the research laboratories to a safe and efficient COVID‐19 molecular diagnostic ward and make available their expertise and time. Importantly, the Board of Directors of the institute, providing encouragement, space and financial room, supported this call fully. Furthermore, the purchasing and safety departments who efficiently sourced personal protection equipment (PPE) for all and provided safe working conditions, critically underpinned the efforts.
The setup of a PCR diagnostic lab
PCR diagnostics requires a quantitative assay, detecting fluorescent signals derived from the amplification of viral genetic material (Figure 2). Methods and equipment are standard in academic labs and the threshold for performing such detection assay is low. The safety measures needed to handle potentially infectious samples were rapidly defined based on the HSM protocols and the WHO biosafety guidelines for handling SARS‐CoV‐2 (see here) and put in place. In only two weeks, 72 samples consisting of nasopharyngeal or oropharyngeal swabs from individuals with suspected COVID‐19 arrived at iMM for analysis. After the initial setup, work was organised in three shifts from 9 a.m. to 9 p.m., in loco or remotely, organized in eight different stations of the pipeline. 1‐ Transport; 2‐ Check in; 3‐ Virus inactivation; 4‐ RNA extraction; 5‐ PCR (all working at IMM) and 6‐ PCR analysis; 7‐ Diagnostic and 8‐ Bridging the Gap (working remotely). The 8th station was to mediate contact on two levels. a) Between the providers of samples and the transport team: to know how many samples were coming and when; how many swabs kits were needed, where and when. b) Between the providers (and their medical doctors that received the PCR reports) and the diagnostic team: to answer any question about eventual repetitions needed, make sure all results were being correctly reported and interpreted as well as to answer any remaining questions.
Figure 2.
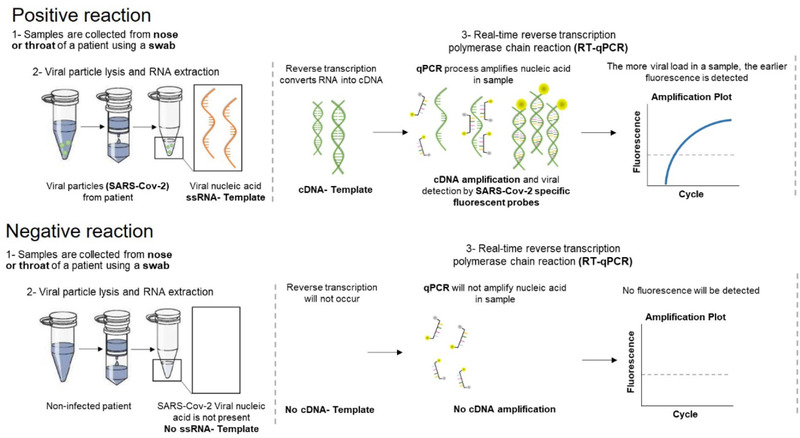
Acute SARS‐CoV‐2 detection via PCR. The schedule highlights the two most likely cases, a positive reaction (top part) when SARS‐CoV‐2 is present or negative reaction (bottom part) when the virus is absent. Detailed protocol available (see here). Schedule credited to Patricia Figueiredo‐Campos and Birte Blankenhaus at iMM.
Three laboratories at iMM were specifically adapted to the SARS‐CoV‐2 detection pipeline – the virus inactivation, RNA extraction and the RT‐PCR laboratories. Upon arrival, barcoded samples were decontaminated, checked‐in and proceed immediately to viral inactivation, manual RNA extraction, and RT‐PCR to quantify SARS‐CoV‐2 RNA. PCR results were analysed and reported remotely. The established pipeline allowed to test up to 900 samples per day. Between April and July, the iMM Diagnostic Task Force performed over 20 000 tests, contributing in an invaluable way to the protection of risk groups, by participating in the screening of COVID‐19 in retirement homes across Portugal.
Due to the rapid spread of the virus and political pressure on governments and the scarcity of resources, boarders started to close, and reagents were hard to obtain. Critical for the success of the operation were the partnerships established very early on with the Portuguese biotech company NZYTech and the HSM. Upon first successful testing of RNA extraction using our in‐house reagents, a reagents supply pipeline was established with NZYTech who formulated a viral RNA extraction kit based on our requirements. Facing the shortage of reagents to perform the molecular test, together we successfully adapted national reagents, the NZY Total RNA isolation kit and the NZY RT‐PCR mix, to the SARS‐Cov‐2 diagnostics.
From setup to validation and reagent sourcing
Academia is full of failed experiments, with largely minor consequences, “trying things out” to see if they work and with constant tinkering of methods. These are not slapdash working practices but a critical part of making progress at the edge of the known and the unknown. It is a stark contrast to the disciplined and methodical practices needed to provide fellow human with important and personal information regarding their health status. A realisation quickly sinking in with all. A robust and well‐defined setup was required.
The protocols for SARS‐CoV‐2 detection were rigorously designed and tested. The first step consisted in validating the RNA extraction protocol with the NZYTech kit using patient samples, in collaboration with the experienced HSM diagnostic labs. Followed by the optimization of the detection of the virus genetic material using the NZY RT‐PCR mix. The PCR protocol adopted was based on that of the US Center for Disease Control and Prevention, using N1 and N2 primer/probe set and literature [1] (see here). The protocol adapted at iMM was validated by the national reference laboratory Instituto Nacional de Saúde Doutor Ricardo Jorge (INSA). Notably, the iMM team kept working in close collaboration with NZYTech that never stop working during the emergency state, producing all the needed reagents and testing the new reagent formulations specifically developed for SARS‐CoV‐2 detection.
Despite the great achievement of setting up a diagnostic laboratory in 2 weeks, testing at iMM could not be enough to cover the country's needs. iMM wrote and made available a detailed Standard Operating Procedures (SOP, see here), encompassing all the laboratory adaptations, safety measures, protocols and procedures put in place in a way that the iMM model could be replicated by any Institute or University willing to join this effort. Nearly 30 research institutes, universities, and polytechnics around Portugal implemented the COVID‐19 diagnostic procedures, most of them following iMM's SOP thereby vastly increasing the country's testing capacity.
The setup of a serology lab
Shortly after the efforts to setup acute SARS‐CoV‐2 diagnostics started, there was another pressing question. How to gain an overview of the viral spread throughout distinct geographical and demographical parts of the country and for front line professions such as healthcare workers? For every infectious agent encountered, our adaptive immune system generates a historical record in the form of memory T and B cells, and circulating antibodies. Antibodies are generated quickly upon infection and can be detected in small blood samples thereby providing a record of past SARS‐CoV‐2 infection. This is used for many infections, such as rubella screening in pregnant women, or screening for vaccination success. At the same time, others throughout the world were thinking the same, with a news story from the Rijksinstituut voor Volksgezondheid en Milieu (RIVM) regarding serology screening of blood donors in the Netherlands drawing our attention (see here).
Similarly to performing PCRs, there is a standard highly sensitive and specific lab‐based method to detect proteins, the enzyme‐linked immunosorbent assay (ELISA). It is the method used by many labs to determine concentrations of proteins. In the case of determining anti‐SARS‐CoV‐2 antibodies, a specific protein from SARS‐CoV‐2 is required so anti‐SARS‐CoV‐2 antibodies are captured and subsequently detected (Figure 3). Plenty of data on coronaviruses (CoV) show that the protein that covers its surface is immunodominant [2, 3]. It was used during the SARS‐CoV and Middle East respiratory syndrome (MERS)‐CoV outbreaks in 2002 and 2012, respectively. At the start of 2020, the SARS‐CoV‐2 sequence was available, but other labs, including the Krammer Lab, US [4] and Bosch lab, NL [5] had already cloned and introduced stabilising mutations and were very generously sharing reagents. Furthermore, at that time a consorted effort of five research institutes in the Lisbon area, named Serology4COVID consortium, was made to help with SARS‐CoV‐2 serological screening. A key member of this consortium was Institute of Experimental Biology and Technology (iBET), who produced the full‐length Spike and receptor binding proteins quickly at high quality and quantity.
Figure 3.
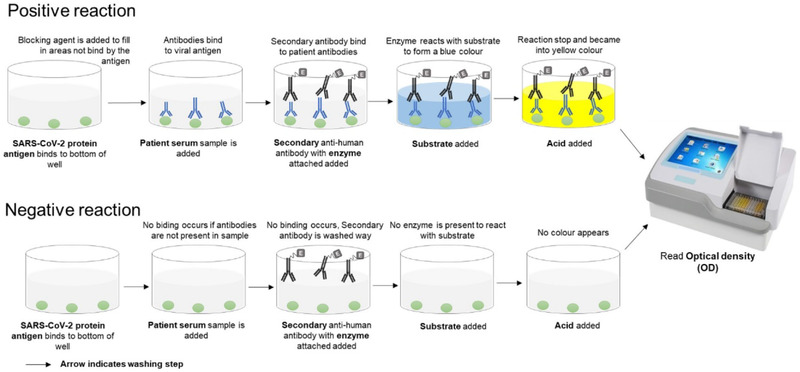
Schedule of the ELISA assay setup. The schedule highlights the two most likely cases, a positive reaction (top part) when SARS‐CoV‐2 was present and antibodies were generated or negative reaction (bottom part) when the virus was not present and no antibodies were made. Detailed protocols available (see here). Schedule credited to Patricia Figueiredo‐Campos and Birte Blankenhaus at iMM.
While the emphasis was on trying to meet demands for acute diagnosis, serology was able to start small. The Veldhoen Lab tissue culture room was rapidly transformed into a BSL2+ room. Most importantly, two volunteers took on the job of setting up and testing a SARS‐CoV‐2 ELISA. There was critical logistical help from the purchasing and safety departments that somehow managed to keep securing PPE for all volunteers despite a growing global shortage. Physicians started to deliver blood samples, of which an additional team of volunteers separated cells and serum. Critically important was the iMM Biobank, with experienced personnel and procedures for handling, storing and tracking samples. They also held a large collection of pre‐COVID‐19 serum samples. The COVID‐19 samples collected through the iMM Biobank can be accessed by researchers worldwide for future research on this disease and constitute an invaluable source of information to further understand this pandemic. As was the case for PCR diagnosis, equipment was present in the institute and repurposed for the serology team.
The most vital part of this effort was the small team of the two volunteers who took it upon themselves the challenge to make the COVID‐19 serology setup a success. Patricia Figueiredo‐Campos, a PhD‐student in the Veldhoen lab, was a research assistant at the Oxford Vaccine Group, UK, as part of preclinical team conducting studies of new and improved vaccines for humans with the setup, testing and improving of ELISAs as main task. She quickly made a comprehensive SOP with all the important steps to setup successfully a sensitive and specific ELISA assay (see here). She got much help and support, while taking care of some of the ordering logistics and health and safety protocols, from Birte Blankenhaus, an experienced postdoctoral researcher in the Veldhoen lab. Importantly, the setup and testing are a two‐person job, with initially serum samples handled with full BSL3 PPE. The small tissue‐culture room, offering space to two biosafety cabinets offered one working place for pipetting serum samples, with sufficient room to work, and a second one containing a plate washer. One‐person pipettes, the second person calls out, monitors the sample and well locations, and helps with any decontamination. Like for an experiment, the conditions for the ELISA were tested systematically to ensure the best results could be obtained [6]. This provided healthcare workers and patients with the certainty that antibodies were present.
However, with the first COVID‐19 wave subduing, people wanted to return to their original projects that had lost very considerable time. A particularly pressing issue in science, where contracts are highly competitive and of short duration. Although reduced cases were reported over the summer, SARS‐CoV‐2 was still very much around and plans were made to undertake large screenings. Yet, we all wanted to be back at the bench and to have our tissue culture room accessible again. How could all the efforts continue without interfering with our academic projects?
Beyond the setup phase
Repurposing of equipment, space and people worked very well as an impulse to setup COVID‐19 diagnostics. Combined academic efforts placing Portugal at the front of COVID‐19 testing per population size [7]. This could not be sustained for long, jeopardising projects and careers as well as institute funds. The Portuguese government, via the Portuguese science foundation (Fundação para a Ciência e a Tecnologia, FCT), provided important project funding for COVID‐19 work in two funding rounds (research4COVID19). We received funding for the PCR setup, the serology setup and for testing neutralising antibodies. This help was very important to recover some of the often‐pre‐financed work undertaken and gratefully received. However, it could not sustain the setup of dedicated diagnostic labs, requiring space, equipment and personnel.
Like most European countries, Portugal does not have a strong history of private donations to scientific institutions. However, a pandemic can be a game changer. The success of iMM leading diagnostic efforts had not gone unnoticed and several organizations offered help to maintain and expand these efforts. The nearly 150 volunteers involved, who created the foundations in which the subsequent efforts were based on, were not forgotten. They were provided with meals and transportation during their work in the different task forces, vouchers for a department store were provided as a token of appreciation, while iMM highlighted the work with a gallery of the volunteers in one of the main meeting rooms and a street billboard with all their pictures (Figure 1).
Figure 1.

A thank you billboard with many of the iMM volunteers who were instrumental in the setup of the COVID‐19 task forces. With thanks for the iMM communication office, photos credited to Jorge Figanier Castro.
Several private donors join in this collective effort: Fundação Oriente and SEMAPA sponsored equipment, while Fundação Luso‐Americana Desenvolvimento (FLAD) critically funded the extension of the iMM Biobank to store samples and their detailed information as a resource to the global scientific community. Additional efforts were made by launching fundraising campaigns for academic institutions to increase the testing capacity. A major private holding, Sociedade Francisco Manuel dos Santos (SFMS), funded the professionalization of the diagnostic labs, purchasing dedicated equipment, and recruiting committed staff. These joint efforts successfully increased the national COVID‐19 testing capacity. They constituted the basis for a structured response to the pandemic, professionalising efforts centralised in a new diagnostic and research centre in Lisbon (see here 1 and 2). The new centre is home to a large network of projects intertwined with the needs of the country and planed according to the evolving pandemic situation. The setup has allowed the dedicated processing of large numbers of swabs by PCR (1500/day, expanding to 3000/day). Many ELISA runs are performed daily, including the testing of over 2500 members of staff, continued testing of COVID‐19 patients, contributions to a large national survey screening, support for scientific projects to determine antibody titres in defined patient cohorts and important work to screen for potential plasma donors for use in convalescent serum therapy with the Portuguese blood bank.
The interactions between the public, private, social and academic sectors have been key for the successful response that a small European country, such as Portugal, has produced in response to the COVID‐19 pandemic. The challenging and unprecedented situation brought visibility to science and to the scientific community that demonstrated extraordinary plasticity and ability to respond. Social recognition, participation in decision‐making and private donations are hopefully some of the long‐term positive consequences that this unfortunate pandemic has brought to our society.
Acknowledgements
We would like to thank all volunteers who helped with this collective effort. We like to acknowledge the funding from the European Union H2020 ERA project (No 667824 – EXCELLtoINNOV) and the Fundação para a Ciência e a Tecnologia (FCT) research4COVID19 (n° 231_596873172, Generating SARS‐CoV2 seroconversion assay: n° 162_596842560, Test, Test, Test: Diagnostic of COVID‐19 and n° 729, High‐throughput SARS‐CoV2 neutralising antibodies assessment).
References
- 1. Nalla, A. K. et al., J Clin Microbiol 2020. 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bosch, B. J. et al., J Virol 2003. 77: 8801‐8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchholz, U. J. et al., Proc Natl Acad Sci U S A 2004. 101: 9804‐9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stadlbauer, D. et al., Curr Protoc Microbiol 2020. 57: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nisreen, M. A. O. et al., Emerging Infectious Disease journal 2020. 26. [Google Scholar]
- 6. Figueiredo‐Campos, P. et al., Eur J Immunol 2020. [Google Scholar]
- 7. Triunfol, M. , Lancet Infect Dis 2020. 20: 783. [DOI] [PMC free article] [PubMed] [Google Scholar]


