Abstract
BACKGROUND
Despite consensus guidelines, concern about severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) transmission has dissuaded patients with cancer from seeking medical care. Studies have shown that contaminated surfaces may contain viable virus for up to 72 hours in laboratory settings. The purpose of this study was to investigate contamination of SARS‐CoV‐2 on commonly used environmental surfaces in a tertiary cancer care center.
METHODS
This study evaluated the incidence of SARS‐CoV‐2 viral RNA in high‐touch outpatient and inpatient cancer center spaces. Surfaces were tested over a 2‐week period after patient or staff exposure but before scheduled disinfection services according to the World Health Organization protocols for coronavirus disease 2019 (COVID‐19) surface sampling. Samples were analyzed via reverse transcriptase–polymerase chain reaction for the presence of SARS‐CoV‐2 RNA.
RESULTS
Two hundred four environmental samples were obtained from inpatient and outpatient oncology clinics and infusion suites, and they were categorized as 1) public areas, 2) staff areas, or 3) medical equipment. One hundred thirty surfaces from 2 outpatient hematology and oncology clinics and 36 surfaces from an inpatient leukemia/lymphoma/chimeric antigen receptor T‐cell unit were examined, and all 166 samples were negative for SARS‐CoV‐2. One of 38 samples (2.6%) from COVID‐19+ inpatient units was positive. Altogether, the positive test rate for SARS‐CoV‐2 RNA across all surfaces was 0.5% (1 of 204).
CONCLUSIONS
This prospective, systematic quality assurance investigation of real‐world environmental surfaces, performed in inpatient and outpatient hematology/oncology units, revealed overall negligible detection of SARS‐CoV‐2 RNA when strict mitigation strategies against COVID‐19 transmission were instituted.
LAY SUMMARY
The potential risks of nosocomial infection with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) have deterred patients with cancer from seeking timely care despite consensus guidelines.
This study has found negligible rates of environmental contamination with SARS‐CoV‐2 across a multitude of commonly used surfaces in outpatient and inpatient hematology/oncology settings with adherence to strict infection control protocols.
Keywords: coronavirus disease 2019 (COVID‐19), environmental surface testing, hematologic malignancies, quality improvement, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), solid tumors, viral transmission
Short abstract
There is a low prevalence of surface contamination with severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in outpatient and inpatient hematology and oncology settings where patients with cancer and COVID‐19 are treated. Although surfaces reveal overall negligible detection of SARS‐CoV‐2 RNA, continued studies are needed to monitor rates of virus transmission and the environmental factors involved in the propagation of the SARS‐CoV‐2 infection.
Introduction
As of August 7, 2020, there were 19 million confirmed cases of coronavirus disease 2019 (COVID‐19) globally, including more than 700,000 deaths related to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), with exponentially increasing numbers. 1 Current evidence suggests that COVID‐19–related morbidity and mortality may be higher in patients with cancer, 2 in part because of underlying comorbidities, increased visits to health care facilities (eg, doctor visits, phlebotomy, imaging, social work, financial consultations, and therapy administration), and inherent or treatment‐induced immunosuppression. 3
The reported fatality rate for patients with cancer harboring SARS‐CoV‐2 is 26% in North America and Spain. 4 Because cancer care provisions have been upended by the COVID‐19 pandemic, consensus guidelines have been issued to help resource‐limited oncology care systems balance reduction of viral exposure while prioritizing cancer care. 5 , 6 , 7 In addition, concern about nosocomial SARS‐CoV‐2 infection continues to deter patients from seeking medical care. 8 In comparison with the prepandemic era, cancer screening has decreased, 9 and this may significantly delay the diagnosis of cancer 10 ; thus, the need for data on the potential risks of infection transmission is amplified.
Although COVID‐19 is known to be transmitted person to person via respiratory droplets 11 and fecal‐oral transmission, 12 contact with environmental surfaces (eg, plastics, metals, and cardboard), on which SARS‐CoV‐2 may be viable for up to 72 hours under laboratory conditions, is also a potential vector of infectivity. Prior studies suggested a role of environmental contamination in health care facilities treating known COVID‐19–infected patients. 13 However, the virus's viability may differ in real‐world clinical environments that promote strict use of personal protective equipment, social distancing, hand hygiene, and disinfectant protocols. 14 To evaluate the potential impact of SARS‐CoV‐2 transmissibility vis‐à‐vis real‐world surfaces in the cancer setting, we assessed surface contamination in multiple outpatient and inpatient hematology/oncology settings that followed strict COVID‐19 risk mitigation strategies. The goal of this prospective quality assurance study was to evaluate the frequency of SARS‐CoV‐2 on a multitude of environmental surfaces in a large tertiary cancer center in New Jersey, which was one of the initial regional epicenters of this worldwide pandemic.
Materials and Methods
Institutional review board approval was obtained from Rutgers University to conduct the study at a COVID‐19 referral center in 2 large and freestanding outpatient cancer clinic suites (ie, malignant hematology and medical oncology), each containing associated infusion centers, and in 2 separate inpatient units (ie, leukemia and lymphoma chimeric antigen receptor T‐cell unit and active COVID‐19 floors, the latter housing patients with cancer actively infected with SARS‐CoV‐2 and persons under investigation [PUIs] for infection). Surface testing for viral RNA in the outpatient infusion suites included spaces where several patients with recent SARS‐CoV‐2 infections were receiving cancer treatment. High‐impact areas were selected on the basis of frequency of use, patient and health care provider contact, and risk of contamination from COVID‐19+ subjects and PUIs due to virus transmissibility.
Surfaces were sampled on Mondays, Wednesdays, and Fridays from June 17, 2020, through June 29, 2020, before any scheduled cleaning and disinfection services but after patient or staff use according to World Health Organization protocols for COVID‐19 surface sampling. 15 The samples were obtained from the inpatient leukemia/lymphoma/chimeric antigen receptor T‐cell unit (Fig. 1), inpatient COVID‐19 and PUI units (Fig. 2), an outpatient malignant hematology clinic and infusion center (Fig. 3), and an outpatient medical oncology clinic and infusion center (Fig. 4). Specimens were analyzed on the same day as collection at the hospital laboratory via real‐time reverse transcriptase–polymerase chain reaction analysis. Health care personnel trained in biosafety and bloodborne pathogen management performed swabbing. Biosafety training was administered online and was offered by Rutgers University. A sterile swab with a polyester tip and plastic shaft was primed with universal viral transport medium and then applied to sites of interest. The swab was then placed in a tube containing 2.5 mL of universal viral transport medium. Samples were immediately refrigerated at 4 °C and held for less than 24 hours before they were manually transported on ice to the in‐hospital laboratory for analysis.
Figure 1.
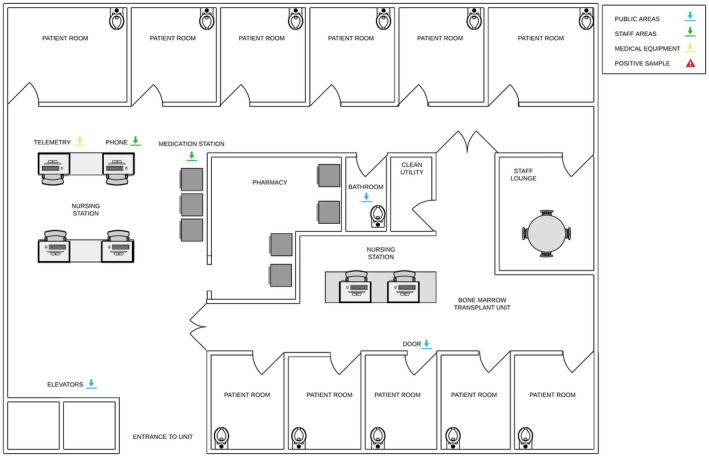
Inpatient leukemia/lymphoma/chimeric antigen receptor T‐cell unit. Samples were taken from the inpatient leukemia/lymphoma/chimeric antigen receptor T‐cell unit in all the locations indicated with arrows (ie, telemetry mouse, phone, medication station, bathroom toilet bowl, elevator buttons, and door handles).
Figure 2.
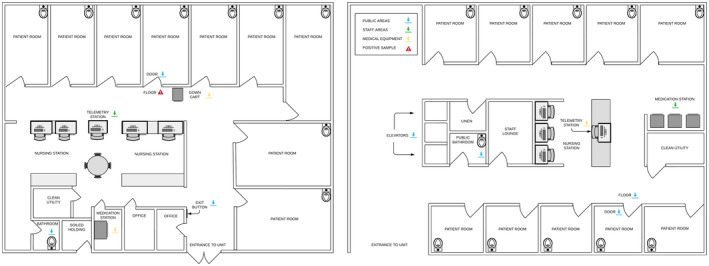
Inpatient COVID‐19/PUI units. Samples were taken from the inpatient COVID‐19/PUI units in all the locations indicated with arrows (ie, floor outside the patient room, door handle, gown cart, telemetry mouse, bathroom toilet bowl, medication station, elevator button, and public bathroom toilet bowel). COVID‐19 indicates coronavirus disease 2019; PUI, person under investigation
Figure 3.
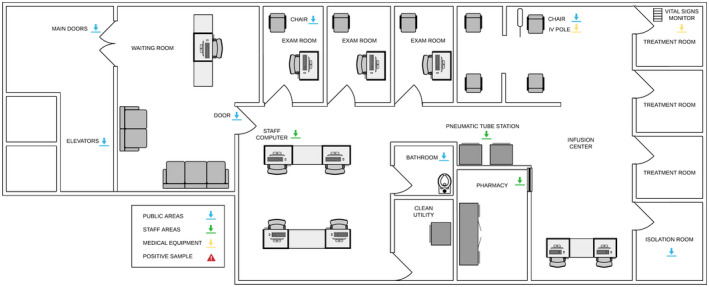
Outpatient malignant hematology. Samples were taken from the outpatient medical hematology clinic and infusion center in all the locations indicated with arrows (ie, door handles, elevator button, staff computer mouse, patient infusion chair, IV pole, bathroom toilet bowl, pneumatic tube station, pharmacy counter, vital signs monitor, and coronavirus disease 2019–positive patient isolation room). IV indicates intravenous.
Figure 4.
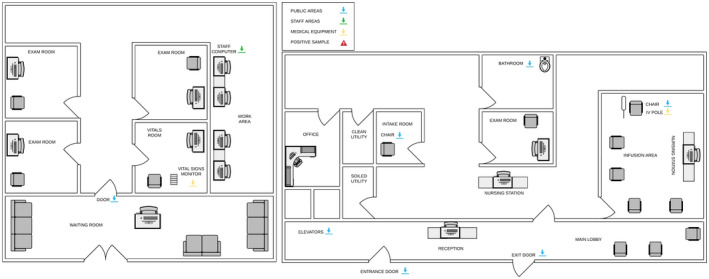
Outpatient medical oncology. Samples were taken from the outpatient medical oncology clinic and infusion center in all the locations indicated with arrows (vital signs monitor, door handle, staff computer mouse, patient intake room chair, elevator button, entrance door handle, exit door handle, bathroom toilet bowl, patient infusion chair, and IV pole). IV indicates intravenous.
The Cobas SARS‐CoV‐2 assay on the Cobas 6800 system, a real‐time reverse transcriptase–polymerase chain reaction assay that allows for sensitive qualitative detection of SARS‐CoV‐2 RNA specimens, was used to analyze the samples. 16 The Cobas SARS‐CoV‐2 assay was granted emergency use authorization on March 12, 2020, for the detection of SARS‐CoV‐2 RNA in nasal, nasopharyngeal, and oropharyngeal swab samples in patients. 17 During analysis, a minimum of 0.6 mL of the sample is transferred into a second container and loaded onto the Cobas 6800 system. The samples are then processed according to the package insert. During the analysis, the samples are combined with probes to ORF1a/b, a nonstructural region that is specific for SARS‐CoV‐2, and envelope E gene, a structural protein shared by the Sarbecovirus subgenus. The Cobas 6800 system adds an internal control to all samples, which is extracted and amplified to control for efficient polymerase chain reaction amplification. Results were qualitatively categorized as 1) positive (2019 novel coronavirus target nucleic acid detected), 2) negative (2019 novel coronavirus target nucleic acid not detected), or 3) invalid (2019 novel coronavirus target nucleic acid cannot be determined). There were no invalid results in our investigation. Internal result validity was determined with Cobas 6800 software on the basis of negative and positive control performance.
Overall, 204 total environmental samples were collected over the study period (Table 1). Testing sites were categorized as 1) public areas (waiting rooms, infusion areas, bathrooms, floors, elevator banks, doors, and examination rooms [including designated isolation rooms for known COVID‐19+ patients/PUIs]), 2) staff areas (computer equipment, pneumatic tubing stations, the pharmacy, and medication rooms), or 3) medical equipment (intravenous poles, chemotherapy bags, vital signs monitors, telemetry stations, and linen carts; see Figs. 1, 2, 3, 4). Please also see the institutional COVID‐19 infection control protocols (Fig. 5) and the supporting information for expanded details on study testing.
TABLE 1.
SARS‐CoV‐2 Surface Testing in Outpatient and Inpatient Cancer Units
| Location | No. Positive | |||
|---|---|---|---|---|
| Total | Patient/Public | Staff | Medical Equipment | |
| Outpatient | 130 | 0/85 | 0/22 | 0/23 |
| Hematologic malignancies | 73 | 0/44 | 0/16 | 0/13 |
| Medical oncology | 57 | 0/41 | 0/6 | 0/10 |
| Inpatient | 74 | 1/42 | 0/18 | 0/14 |
| Lymphoma/leukemia/CAR T cell | 36 | 0/18 | 0/12 | 0/6 |
| COVID‐19/PUI unit | 38 | 1/24 a | 0/6 | 0/8 |
Abbreviations: CAR, chimeric antigen receptor; COVID‐19, coronavirus disease 2019; PUI, person under investigation; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Patient/public areas included toilets, waiting rooms, elevator banks, patient examination rooms (including designated isolation rooms for COVID‐19+ patients/PUIs), infusion chairs, clinic entrances/exits, and floors. Staff areas included pneumatic tube stations, the nursing station, the pharmacy, and computer equipment (ie, mouses and keyboards). Medical equipment included vital signs monitors, telemetry stations, intravenous poles, and linen carts.
The 1 positive result came from the floor with a room with a patient with multiple medical comorbidities on day 9 of hospitalization who was admitted with COVID‐19 pneumonia.
Figure 5.
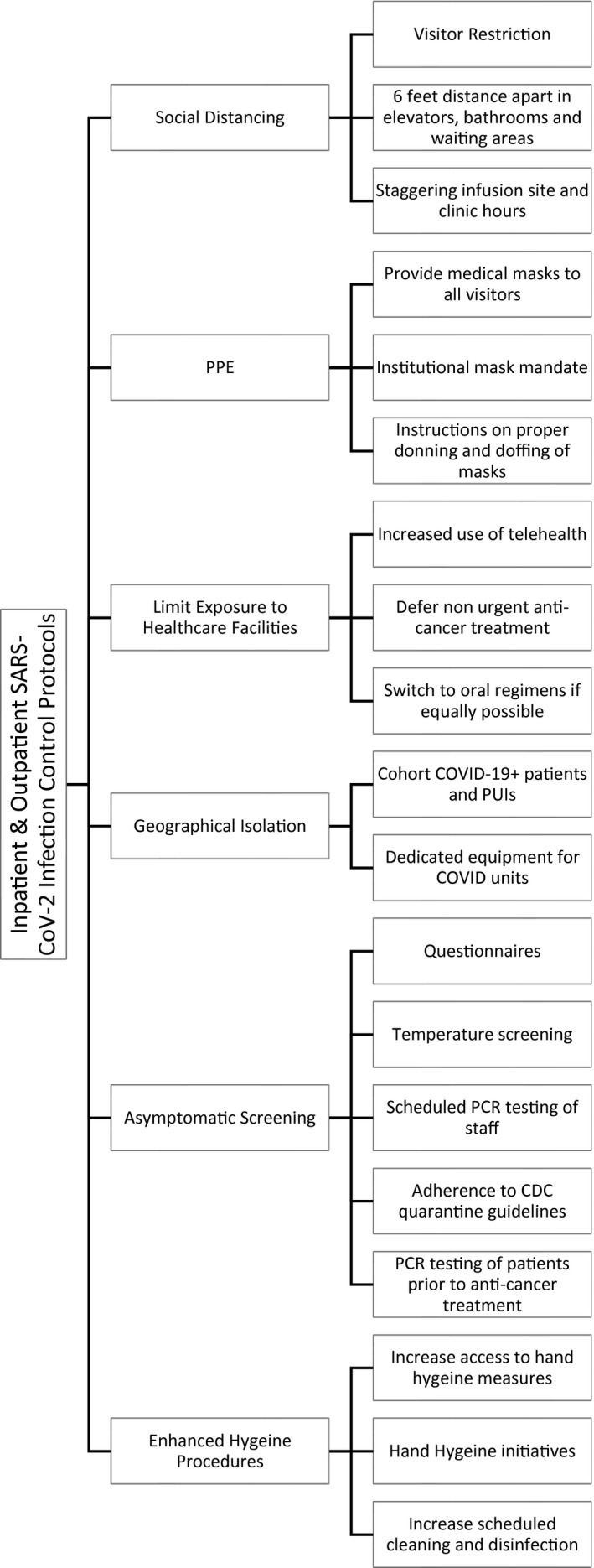
Inpatient and outpatient SARS‐CoV‐2 infection control policies. Inpatient and outpatient SARS‐CoV‐2 infection control policies included the following: 1) visitor restriction (visitors were prohibited from the treatment areas or clinic unless they are needed for patient safety and patient care as preapproved by the nursing manager), 2) defermentof nonurgent anticancer treatment (testing all patients suspected of having COVID and deferring anticancer treatment if possible until test results arrive), 3) an institutional mask mandate (all visitors, adult/pediatric patients, and staff are required to wear a surgical mask upon entry to the facility), 4) staggering infusion site and clinic hours (clinic hours and infusion hours were not held on overlapping times but instead were given dedicated time slots), 5) increased use of telehealth (increased use of telemedicine visits when appropriate; when there is high suspicion for SARS‐CoV‐2 infection, then the patient's appointment is rescheduled or converted to a telemedicine visit as appropriate), 6) scheduled polymerase chain reaction testing of staff (all staff must undergo SARS‐CoV‐2 polymerase chain reaction testing at scheduled intervals and adhere to appropriate CDC guidelines on the basis of test results and symptoms), 7) questionnaires (screening all staff and adult/pediatric patients before they enter inpatient or outpatient facilities for infectious symptoms, travel history, and regular antipyretic use), 8) temperature screening (temperature checks of all staff and adult/pediatric patients before they enter any health care facility), 9) hand hygiene initiatives (encouraging staff to enhance sanitation of high‐traffic areas and practice good hygiene techniques), and 10) increased access to hand hygiene measures (increasing the availability and enforcing the use of hand sanitization dispensers near high‐use areas [elevators, doors, and handles]). CDC indicates Centers for Disease Control and Prevention; COVID, coronavirus disease; COVID‐19, coronavirus disease 2019; PCR, polymerase chain reaction; PPE, personal protective equipment; PUI, person under investigation; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Results
Among the 130 surfaces examined from 2 outpatient hematology/oncology clinics and the 36 samples tested from the inpatient leukemia/lymphoma/chimeric antigen receptor T‐cell units, all 166 surfaces were negative for SARS‐CoV‐2 viral RNA. Among the inpatient COVID‐19+ and PUI units, there was 1 positive (surface area) out of 38 samples (2.6%). The 1 positive sample was taken from the floor of an elderly patient with multiple medical comorbidities who was treated with remdesivir, dexamethasone, and apixaban for COVID‐19 pneumonia. Collectively, the positive test rate for SARS‐CoV‐2 RNA across all surfaces in the outpatient/inpatient units was 0.5% (1 of 204).
Discussion
This prospective, systematic quality assurance investigation of real‐world environmental surfaces, performed in inpatient and outpatient hematology/oncology units, revealed overall negligible detection of SARS‐CoV‐2 RNA. These data highlight several important points, including the overall adequacy of current, detailed infection control policies in limiting environmental surface contamination with SARS‐CoV‐2. These policies include increased use of telephone screening; nursing‐directed triage for PUIs/COVID‐19+ patients; visitor restrictions; telehealth; and the importance of personal protective equipment, social distancing, and disinfection protocols as outlined in Figure 5.
In addition, the 1 positive sample from a COVID‐19 unit reinforces the importance of physical separation of patients infected with SARS‐CoV‐2. We speculate that the floor may have contained fomites composed of viral RNA. Our finding is similar to a study from Singapore that screened surface and air samples from hospital rooms of COVID‐19 patients for SARS‐CoV‐2 RNA and found that the floor was most likely to be contaminated. 18 Moreover, multiple studies have established the inverse relationship of viral burden with increased time after symptom onset, 19 and this may also explain the low levels of viral detection.
Early research into the surface stability of SARS‐CoV‐2 20 raised concerns about fomite transmission as a major contributor to spread of the disease. Significant environmental contamination with SARS‐CoV‐2 RNA was found in various areas in hospital wards and on commonly touched surfaces in health care facilities in China and Singapore where patients with COVID‐19 were treated. 21 , 22 Although multiple studies have found SARS‐CoV‐2 on environmental surfaces, 18 , 22 , 23 , 24 few have attempted to culture the virus from positive samples. Despite the uncertainty about whether viable virus exists on these positive samples, respiratory droplets can dwell on surfaces and can theoretically infect an individual through touching of the eyes, nose, and mouth, although this mode of transmission is likely less dominant, especially in settings with standardized cleaning and screening protocols. In the context of potentially immunocompromised patients with cancer, it is important to understand the role of environmental contamination to further protect this vulnerable population. Despite infection control guidelines specific to oncology care, 25 cancer‐related encounters have remained below expectant volumes, 26 and this perhaps reflects persistent concerns about acquiring a nosocomial COVID‐19 infection.
Because the focus of this investigation was to evaluate surface contamination, we did not evaluate airborne transmission. However, potential airborne diffusion of the virus has been investigated in China, 27 , 28 and the virus detection rate remained low when the application of more rigorous disinfection and infection control procedures were followed. Other limitations included the inability to analyze the complete surface area of the location, which may have reduced sensitivity. Additionally, we did not attempt to culture SARS‐CoV‐2 from our positive sample; it is unknown if it contained live virus.
Altogether, we demonstrated that SARS‐CoV‐2 presence on environmental surfaces as detected by PCR in clinical cancer units was extremely low, especially in the outpatient setting. The results of this study are reassuring and should reduce concerns for patients and health care providers about infection transmission from environmental surfaces in outpatient and inpatient oncology spaces when strict mitigation strategies against SARS‐CoV‐2 transmission are instituted. Continued studies are needed to monitor rates of virus transmission and the environmental factors involved in the propagation of the SARS‐CoV‐2 infection.
Funding Support
No specific funding was disclosed.
Conflict of Interest Disclosures
Andrew M. Evens has received honoraria for research advisory boards from Seattle Genetics, MorphoSys, Mylteni, Epizyme, Novartis, Karyopharm, Pharmacyclics, and AbbVie. The other authors made no disclosures.
Author Contributions
Mansi R. Shah: Conception and planning of the study; data collection; creation of tables and figures; writing of the manuscript; critical feedback; and shaping of the research, analysis, and manuscript. Imraan Jan: Conception and planning of the study; critical feedback; and shaping of the research, analysis, and manuscript. Jeremy Johns: Data collection; creation of tables and figures; critical feedback; and shaping of the research, analysis, and manuscript. Kuldip Singh: Data collection; creation of tables and figures; critical feedback; and shaping of the research, analysis, and manuscript. Pallavi Kumar: Data collection; creation of tables and figures; critical feedback; and shaping of the research, analysis, and manuscript. Norma Belarmino: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Kara J. Saggiomo: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Carolyn Hayes: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Kimyatta Washington: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Deborah L. Toppmeyer: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Bruce G. Haffty: Study design; critical feedback; and shaping of the research, analysis, and manuscript. Steven K. Libutti: Critical feedback and shaping of the research, analysis, and manuscript. Andrew M. Evens: Conception and planning of the study; writing of the manuscript; critical feedback; and shaping of the research, analysis, and manuscript.
Supporting information
Supplementary Material
Shah MR, Jan I, Johns J, Singh K, Kumar P, Belarmino N, Saggiomo KJ, Hayes C, Washington K, Toppmeyer DL, Haffty BG, Libutti SK, Evens AM. SARS‐CoV‐2 nosocomial infection: Real‐world results of environmental surface testing from a large tertiary cancer center. Cancer. 2021. 10.1002/cncr.33453
We acknowledge Jonathan Bodnar (Laboratory Director, Robert Wood Johnson University Hospital) for providing testing kits and in‐house laboratory analysis.
References
- 1. World Health Organization . WHO coronavirus disease (COVID‐19) dashboard. Accessed July 20, 2020. https://covid19.who.int/
- 2. Dai M, Liu D, Liu M, et al. Patients with cancer appear more vulnerable to SARS‐CoV‐2: a multicenter study during the COVID‐19 outbreak. Cancer Discov. 2020;10:783‐791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang L, Zhu F, Xie L, et al. Clinical characteristics of COVID‐19–infected cancer patients: a retrospective case study in three hospitals within Wuhan, China. Ann Oncol. 2020;31:894‐901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuderer NM, Choueiri TK, Shah DP, et al. Clinical impact of COVID‐19 on patients with cancer (CCC19): a cohort study. Lancet. 2020;395:1907‐1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hanna TP, Evans GA, Booth CM. Cancer, COVID‐19 and the precautionary principle: prioritizing treatment during a global pandemic. Nat Rev Clin Oncol. 2020;17:268‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al‐Shamsi HO, Alhazzani W, Alhuraiji A, et al. A practical approach to the management of cancer patients during the novel coronavirus disease 2019 (COVID‐19) pandemic: an international collaborative group. Oncologist. 2020;25:e936‐e945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Richards M, Anderson M, Carter P, Ebert BL, Mossialos E. The impact of the COVID‐19 pandemic on cancer care. Nat Cancer. 2020;1:565‐567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartnett KP. Impact of the COVID‐19 pandemic on emergency department visits—United States, January 1, 2019–May 30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69:699‐704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burki TK. Cancer guidelines during the COVID‐19 pandemic. Lancet Oncol. 2020;21:629‐630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaufman HW, Chen Z, Niles J, Fesko Y. Changes in the number of US patients with newly identified cancer before and during the coronavirus disease 2019 (COVID‐19) pandemic. JAMA Netw Open. 2020;3:e2017267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yeo C, Kaushal S, Yeo D. Enteric involvement of coronaviruses: is faecal‐oral transmission of SARS‐CoV‐2 possible? Lancet Gastroenterol Hepatol. 2020;5:335‐337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu J, Ouyang W, Chua ML, Xie C. SARS‐CoV‐2 transmission in patients with cancer at a tertiary care hospital in Wuhan, China. JAMA Oncol. 2020;6:1108‐1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu DK, Akl EA, Duda S, et al. Physical distancing, face masks, and eye protection to prevent person‐to‐person transmission of SARS‐CoV‐2 and COVID‐19: a systematic review and meta‐analysis. Lancet. 2020;395:1973‐1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. World Health Organization . Surface Sampling of Coronavirus Disease (COVID‐19): A Practical “How To” Protocol for Health Care and Public Health Professionals. World Health Organization; 2020. [Google Scholar]
- 16. US Food and Drug Administration . Cobas® RU. SARS‐CoV‐2: package insert. Accessed December 5, 2020. https://www.fda.gov/media/136049/download
- 17. Poljak M, Korva M, Knap Gašper N, et al. Clinical evaluation of the Cobas SARS‐CoV‐2 test and a diagnostic platform switch during 48 hours in the midst of the COVID‐19 pandemic. J Clin Microbiol. 2020;58:e00599‐e00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by SARS‐CoV‐2 in hospital rooms of infected patients. Nat Commun. 2020;11:2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26:672‐675. [DOI] [PubMed] [Google Scholar]
- 20. Van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382:1564‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye G, Lin H, Chen L, et al. Environmental contamination of the SARS‐CoV‐2 in healthcare premises: an urgent call for protection for healthcare workers. medRxiv. Preprint posted online March 16, 2020. doi: 10.1101/2020.03.11.20034546 [DOI] [Google Scholar]
- 22. Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) from a symptomatic patient. JAMA. 2020;323:1610‐1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yung CF, Kam KQ, Wong MSY, et al. Environment and personal protective equipment tests for SARS‐CoV‐2 in the isolation room of an infant with infection. Ann Intern Med. 2020;173:240‐242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ye G, Lin H, Chen S, et al. Environmental contamination of SARS‐CoV‐2 in healthcare premises. J Infect. 2020;81:e1‐e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van de Haar J, Hoes LR, Coles CE, et al. Caring for patients with cancer in the COVID‐19 era. Nat Med. 2020;26:665‐671. [DOI] [PubMed] [Google Scholar]
- 26. London JW, Fazio‐Eynullayeva E, Palchuk MB, Sankey P, McNair C. Effects of the COVID‐19 pandemic on cancer‐related patient encounters. JCO Clin Cancer Inform. 2020;4:657‐665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Ning Z, Chen Y, et al. Aerodynamic analysis of SARS‐CoV‐2 in two Wuhan hospitals. Nature. 2020;582:557‐560. [DOI] [PubMed] [Google Scholar]
- 28. Li YH, Fan YZ, Jiang L, Wang HB. Aerosol and environmental surface monitoring for SARS‐CoV‐2 RNA in a designated hospital for severe COVID‐19 patients. Epidemiol Infect. 2020;148:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


