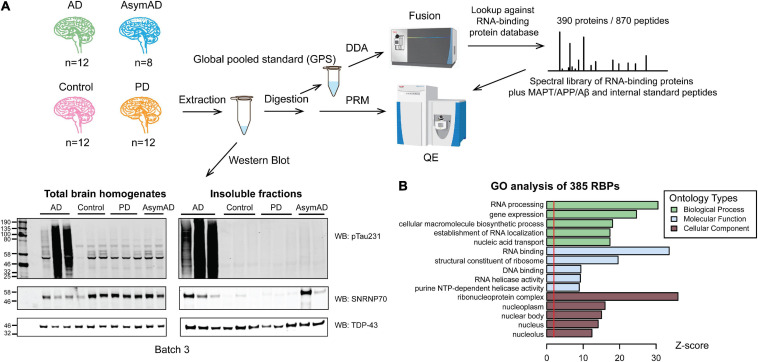
FIGURE 1.
Workflow of the targeted PRM method for quantification of RNA-binding proteins in detergent-insoluble fractions of human brains. (A) Workflow beginning with dorsolateral prefrontal cortex brain samples from control group (n = 12), Alzheimer’s disease (AD, n = 12), Asymptomatic AD (AsymAD, n = 8), and Parkinson’s disease (PD, n = 12) from which insoluble fractions were extracted and digested for subsequent quantification. Western blotting total homogenates and insoluble fractions of each group (n = 3, each) for TPD-43 (loading control), phosphorylated tau (pThr231-tau) and U1-70K indicates successful insoluble fraction extraction. A global pooled standard (GPS) was deeply profiled to identify the total insoluble proteome using an Orbitrap Fusion mass spectrometer. A custom targeted RNA-binding protein (RBP) peptide list and corresponding spectra library was designed for PRM, including 870 peptides from 385 RBPs plus tau, Aβ, APP and internal standard peptides. Finally, PRM was run on Q-Exactive mass spectrometer for all 44 samples using 6 GPS samples to normalize between batches. (B) GO analysis of detected 385 RBPs linked proteins to “RNA processing” and “RNA binding” biological processes and molecular functions, respectively.