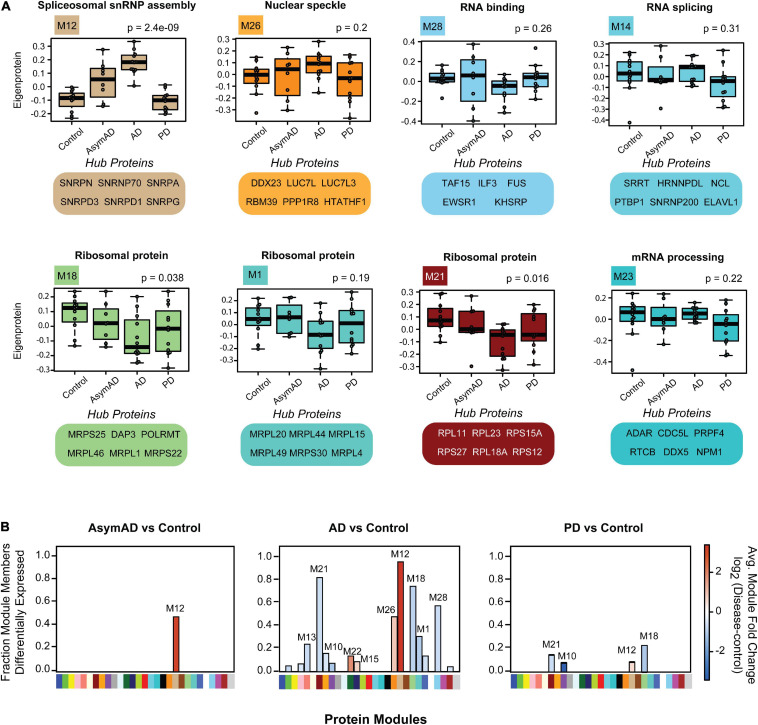
FIGURE 5.
Module-wise insolubility changes according to disease status. (A) A highlight of modules that were significantly correlated with disease groups. Top GO terms were highlighted for each module. All of 29 individual modules overrepresent gene ontologies. For example, M12 shows a group of proteins that belong to “spliceosome snRNP assembly;” M26 includes “nuclear speckle” proteins. The top six proteins which drive the module insolubility, called eigenproteins, are listed below each box plot. M12 and M26 showed stage-specific enrichment in detergent-insoluble fraction of AsymAD and AD. Modules M18, M21 and M28 exhibited decreased insolubility in AD. Modules M14 and M23 experience decreased insolubility in PD only. The central horizontal bar in each box is the 50th percentile (median), and hinges of the box extents represent the interquartile range of the two middle quartiles of data within a group. The farthest data points, up to 1.5 times the interquartile range away from box hinges, define the extent of whiskers (error bars). Significance was measured using one-way non-parametric ANOVA, Kruskal-Wallis p-Values. (B) Module eigenprotein levels for each pairwise comparison between disease groups and controls are shown. Direction of solubility change is colored according to log2 fold change colored scale. The percentage of module members that were differentially abundant in the insoluble fraction in disease compared with control is plotted on the y-axis.