Abstract
Diagnostics is crucial for a prompt identification of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infected patients, their isolation and treatment. Real‐time PCR is the reference method for the diagnosis of SARS‐CoV‐2 infection; however, the unprecedented increase in the number of infections worldwide calls for faster and easy methods that do not require skilled personnel and special equipment. Rapid antigen tests have been developed and used as first line screening. Here, we assessed the performance of a rapid antigen test in comparison to a real‐time qualitative PCR as gold standard. Fifty nasopharyngeal swabs from suspected cases of SARS‐CoV‐2 infection have been tested by Coris coronavirus disease 2019 Ag Respi‐Strip test and Allplex 2019n‐CoV assay. Of the 50 nasopharyngeal swabs tested, 11 were negative by both tests, 27 were negative by Ag test but positive by real‐time PCR, and 12 were positive by both methods. PCR detected the 39 positive samples at a median cycle threshold (Ct) value of 22.78 (mean: 24.51; range: 13.59–39.6). In the 12 concordant samples, the median Ct value was 17.37. The sensitivity of the Ag test was 30.77% (95% confidence interval [CI]: 17.02%–47.57%), specificity 100% (95% CI: 71.51%–100.00%), positive predictive value 100%, negative predictive value 85.25% (95% CI: 82.42%–87.69%), and accuracy 86.15% (95% CI: 73.45%–94.28%). The level of agreement between the two tests was poor, k = 0.164. The Ag test performs well in the presence of high viral loads, whereas lower levels are missed. Considering the poor sensitivity of the method, real‐time PCR remains the gold standard as front line screening for SARS‐CoV‐2 infection.
Keywords: COVID‐19, nasopharyngeal swabs, rapid antigen test, real‐time PCR, SARS‐CoV‐2
1. INTRODUCTION
At the end of December 2019, an outbreak of pneumonia of unknown etiology was reported in Wuhan, Hubei province, China. Inoculation of the respiratory secretions of infected individuals into Vero E6 and Huh7 cell lines and human airway epithelial cells brought to the isolation of a novel virus whose genome sequence showed belonging to the Coronaviridae family, subgenus Sarbecovirus. The novel coronavirus is related to the human SARS‐CoV and some bat SARS‐like coronaviruses, and therefore named severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). 1 , 2 SARS‐CoV‐2 is the causative agent of coronavirus disease 2019 (COVID‐19). Efficient human‐to‐human transmission allowed the spread of the novel coronavirus all over the world; thus, on March 11, 2020, World Health Organization (WHO) declared COVID‐19 as a pandemic (WHO Director General/Speeches/Detail).
SARS‐CoV‐2 testing is needed to limit outbreak through identification of infected individuals and for contacts tracing. The ideal test would be no just the one capable of identifying the presence of SARS‐CoV‐2, but the one that is able to identify those individuals that can transmit the infection to others. They are probably those with the highest viral load in the upper respiratory tract, which usually occurs in the first days of infection. 3 Antigen tests have been indicated as the tests that have the potential of quickly identifying individuals that can transmit infectious viral particles. Although less sensitive than molecular tests that detect viral RNA, antigen tests perform well on individuals with high viral load in their upper respiratory tract. 4 , 5 Antigen test is easy to perform, it does not requires skilled personnel and give results in about 15 min.
In this study, we compared the performance of a rapid antigen test, COVID‐19 Ag Respi‐Strip, with that of a real‐time PCR assay, Allplex 2019n‐CoV assay.
2. MATERIALS AND METHODS
2.1. Clinical specimens
Fifty nasopharyngeal swabs collected at the Emergency Department or Infectious Diseases ward of the University Hospital Tor Vergata, Rome, between May and September 2020, were received by the Virology Lab and tested by real‐time PCR and antigen test for the presence of SARS‐CoV‐2. The median, mean, and age range and sex ratio (men/women) of the study population was calculated.
2.2. Detection of SARS‐CoV‐2 by real‐time reverse‐transcriptase PCR
Detection of SARS‐CoV‐2 RNA in nasopharyngeal swabs was performed by real‐time reverse transcriptase (RT) PCR using the Allplex 2019n‐CoV assay, designed for the qualitative detection of SARS‐CoV‐2 in respiratory samples (Seegene). After RNA extraction and PCR set‐up on NIMBUS (Seegene), an automated liquid handling workstation, real‐time RT‐PCR was performed on the CFX96TMDx platform (Bio‐Rad Laboratories, Inc.) followed by interpretation of the results by Seegene's Viewer Software. The Allplex 2019n‐CoV assay is a multiplex real‐time PCR targeting the envelope (E) gene, common to the coronaviruses belonging to the Sarbecovirus subgenus, and the specific nucleocapsid (N) and RNA‐dependent‐RNA‐polymerase (RdRp) genes; complying with the international validated protocols.
The Allplex 2019n‐CoV assay has a limit of detection of 4167 copies/ml and a sensitivity of 100 copies/reaction (EUA‐FDA, Segeene Allplex 2019‐nCoV Assay). Target genes amplified within ≤40 Ct are considered detected.
2.3. Rapid antigen test for the detection of SARS‐CoV‐2
SARS‐CoV‐2 antigen was detected by COVID‐19 Ag Respi‐Strip (Coris BioConcept), following manufacturer's instructions. This assay uses membrane technology based on colloidal gold nanoparticles and monoclonal antibodies directed against a highly conserved antigen of the nucleoprotein of SARS‐CoV and SARS‐CoV‐2. In addition, another monoclonal antibody is conjugated to colloidal gold nanoparticles. The monoclonal antibodies are immobilized onto the nitrocellulose membrane.
One hundred µl of nasopharyngeal secretions are mixed with four drops (about 100 µl) of lysis buffer inside a tube and then the strip is added. The respiratory sample diffuses with the solubilized conjugate coming into contact with anti‐SARS antibody adsorbed onto the strip. In the presence of SARS‐CoV‐2, the conjugate‐SARS‐CoV complex remains bound to the anti‐SARS‐CoV‐2 antibody immobilized onto the nitrocellulose membrane. A red line will develop followed by a second control red line within 15 min.
2.4. Statistics
We described the sample in terms of age (as mean, median and range in both assay), and sex ratio. The performance of Covid‐19 Ag Respi‐Strip was assessed calculating its sensitivity and specificity. Allplex 2019n‐CoV assay was considered as the “gold standard” for evaluating the positivity or negativity to the samples under analysis. The samples resulting positive or negative to the methods tested were considered to be true positive and true negative samples, respectively. The level of agreement between the two assays was evaluated by Cohen's kappa coefficient (κ), assuming as scarce agreement a κ value less than 0.4. All analyses were performed using SPSS v. 22.0.
3. RESULTS
Fifty nasopharygeal swabs from 50 patients with suspected SARS‐CoV‐2 infection were tested by real‐time RT‐PCR and rapid Ag Test. The median age of the study population was 53.5 years, (mean 53.14; range: 15–94 years old). Sex ratio was 0.92 (24 men and 26 women). Eleven samples tested negative by both methods, 27 were positive by PCR but negative by Ag test, and 12 tested positives by both methods, Table 1. Allplex 2019n‐CoV assay detected SARS‐CoV‐2 RNA in 39 out of 50 samples with a median cycle threshold value of 22.78 for all three genes (mean: 24.51; range: 13.59–39.6), Figure 1A. In the 12 nasopharyngeal swabs positive by both methods, the median cycle threshold was 17.37, Figure 1B. The median age of these patients was 47 years (mean 40, range: 15–88 years old); 7 men and 5 women.
Table 1.
Detection of SARS‐CoV‐2 by COVID‐19 Ag Respi‐Strip and Allplex 2019n‐CoV assays
| RT‐PCR | |||
|---|---|---|---|
| Detected | Not detected | ||
| Ag Respi‐Strip | Detected | 12 | 0 |
| Not detected | 27 | 11 | |
Abbreviation: COVID‐19, coronavirus disease 2019; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 1.
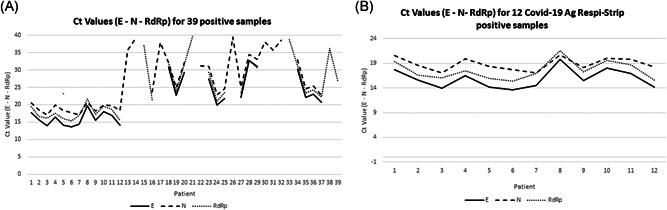
Determination of SARS‐CoV‐2 viral load in the 39 positive nasopharyngeal swabs. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
The sensitivity of the Ag test was 30.77% (95% confidence interval [CI]: 17.02%–47.57%), specificity 100% (95% CI: 71.51%–100.00%). Assuming a prior probability of infection (prevalence) of 20% based on the detection of SARS‐CoV‐2 in the patients admitted to our hospital, the positive predictive value (PPV) was 100%, negative predictive value 85.25% (95% CI: 82.42%–87.69%), and accuracy 86.15% (95% CI: 73.45%–94.28%). The level of agreement between the two tests was poor, k = 0.164.
4. DISCUSSION
A prompt diagnosis of suspected cases of SARS‐CoV‐2 infection is required for properly manage the infected patients as well as to limit the spread of the virus. Antigen tests respond to the need of rapidity, but their performance is conditioned by factors such as the viral load, the quality of the specimen and the processing phase. In this study, the performance of the COVID‐19 Ag Respi‐Strip was compared to that of the Allplex 2019n‐CoV assay, a real‐time PCR assay routinely used in our laboratory. The data obtained suggest that the Ag test performs well in the presence of high viral load in the nasopharyngeal swab corresponding approximately at a median Ct value of 17.37. No antigen was detected when the Ct value was more than 17.37. Actually, the sensitivity of the assay was 30.77% while the specificity was 100%. The sensitivity reported in this study is much lower than that claimed by the manufacturer. This discrepancy might be explained by the use in this study of samples detected positive within a wide range of Ct value, while the manufacturer determined the sensitivity of its assay on samples detected within a Ct less than 25, which contain a higher target concentration, obtaining a higher sensitivity. The PPV and NPV was 100% and 85.25%, respectively, and accuracy 86.15%. Finally, the Cohen's kappa value was 0.14 indicating a poor agreement between the two assays. These data are in line with those published recently by Scohy et al. 5
The poor sensitivity of the Ag tests is well known and it is related to its technical design. Unlike PCR, Ag tests do not amplify their signal so low amount of target protein can be missed. For this reasons, WHO recommends rapid antigen tests that have a sensitivity of ≥80% and a specificity of ≥97%, while ECDC suggests using tests with ≥90% sensitivity and ≥97% specificity, features close to those of a real‐time PCR assay. 6 In the absence of Ag test with the performances recommended by international agencies, it is advisable not to use the Ag test as the sole screening tool for the diagnosis of SARS‐CoV‐2 infection. Negative cases should be confirmed by real‐time PCR that remains the gold standard for detecting SARS‐CoV‐2.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Marco Ciotti conceived the study and wrote the manuscript; Massimo Maurici performed the statistical analysis; Massimo Pieri performed the experiments; Massimo Andreoni and Sergio Bernardini, supervised the study and critically revised the manuscript.
Ciotti M, Maurici M, Pieri M, Andreoni M, Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS‐CoV‐2 infection. J Med Virol. 2021;93:2988–2991. 10.1002/jmv.26830
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 4. Manabe YC, Sharfstein JS, Armstrong K. The need for more and better testing for COVID‐19. JAMA. Published online November 13 2020;324:2153. https://jamanetwork.com/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scohy A, Anantharajah A, Bodéus M, Kabamba‐Mukadi B, Verroken A, Rodriguez‐Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID‐19 diagnosis. J Clin Virol. 2020;129:104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. https://www.ecdc.europa.eu/sites/default/files/documents/Options-use-of-rapid-antigen-tests-for-COVID-19.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


