Fig. 1.
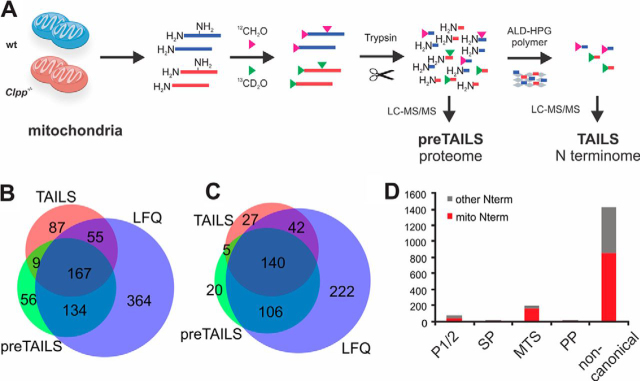
Mouse heart N terminome profiling.A, Scheme of the TAILS workflow. Protein N termini and Lys side chains from wild type (wt) and Clpp−/− mitochondrial proteins were labeled with light and heavy formaldehyde, respectively, pooled and digested with trypsin. A “preTAILS” aliquot was withdrawn for labeling control and determination of protein abundance changes. In the next reaction, peptides with unlabeled N termini resulting from tryptic digestion were covalently captured with a high-molecular weight aldehyde-functionalized ALD-HPG polymer. Removal of the polymer with bound peptides by ultrafiltration left labeled N-terminal peptides highly enriched in the flow-through for LC-MS/MS analysis. B, Overlap of all proteins identified by the preTAILS and TAILS in this analysis and our previous LFQ proteome analysis (13). C, Overlap of proteins with UniProt-annotated mitochondrial localization between preTAILS, TAILS and LFQ data sets. D, Positional annotation classifying protein N-terminal peptides identified after TAILS enrichment into 5 categories, those matching position 1 or 2 of the protein model, matching within 5 amino acids of annotated or predicted signal peptide (SP), mitochondrial targeting signal (MTS) cleavage sites, propeptide maturation sites (PP) or those matching to “unexpected” positions within the protein model. Red bars indicate proteins with mitochondrial location as annotated by UniProt.