Fig. 2.
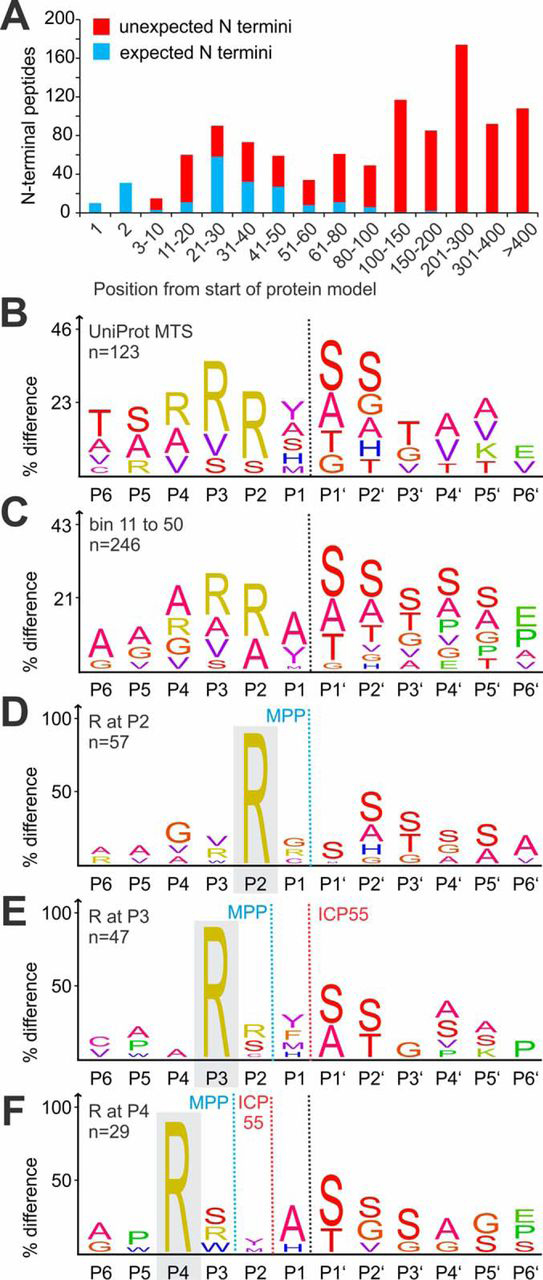
Analysis of the mitochondrial mouse heart N terminome. A, Start position of the 1058 identified N-terminal peptides in relation to the corresponding protein model. Blue indicates expected termini mapping to positions 1 or 2 or within 5 residues from a UniProt-annotated mitochondrial targeting signal (MTS) cleavage site, not annotated unexpected N termini are shown in red. IceLogos show amino acids overrepresented in (B) 123 unique cleavage sites matching within 5 aa of annotated MTS cleavage sites and (C) 246 unique cleavage sites derived from peptides mapping to positions >10 and <51. Further iceLogos visualize amino acids overrepresented among (D) 57 cleavages sites with Arg at P2, (E) 47 cleavage sites with Arg at P3, and (F) 29 cleavage sites with Arg at P4. The dashed black line indicates start of experimentally determined N termini, dashed blue line indicates putative MPP cleavage site, dashed red line putative ICP55 cleavage site.
