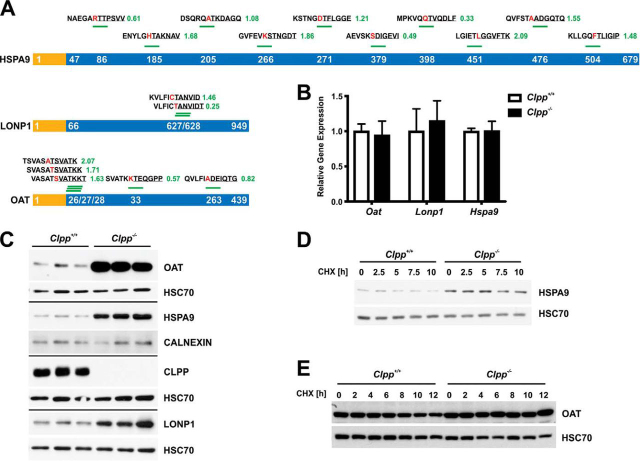
Fig. 4.
Validation of candidate ClpXP substrates identified by TAILS.A, Putative ClpXP substrates with increased N termini abundance. MTS (yellow) and mature proteins (blue) are shown with the starting positions of the accumulating N termini indicated by white numbers. Above the proteins, cleavage windows are stated with the difference in abundance (log2(Clpp−/−/wt)). Amino acids at the P1 position preceding the cleavage site are highlighted in red, the detected peptide sequence is underlined. B, Western blots of steady state protein levels in heart lysates. HSC70 and CALNEXIN were used as loading controls for the respective blots. C, Relative gene expression with qPCR of Oat, Lonp1 and Hspa9. D, CHX chase experiment of HSPA9 in MEFs. HSC70 was used as loading control. E, CHX chase experiment of OAT in MEFs. HSC70 was used as loading control.