Fig. 5.
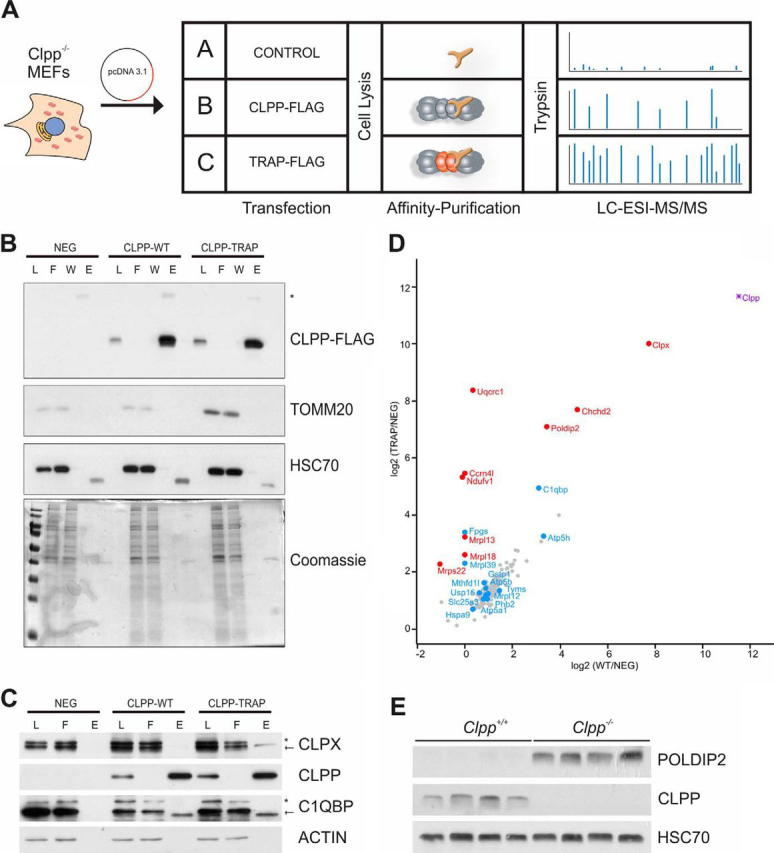
Identification of candidate ClpXP substrates by trapping.A, Trapping workflow: Control, CLPP-WT and CLPP-TRAP containing plasmids were transfected into Clpp−/− MEFs. Cells were lysed and FLAG-tagged CLPP was affinity purified with magnetic beads. CLPP and bound proteins were eluted from the beads, subjected to trypsin digestion and quantified with LC-MS. B, Western blotting and Coomassie-stained gel of total lysate (L), flow-through (F), washing (W) and elution (E) fractions. TOMM20 and HSC70 were used as controls for unspecific mitochondrial and cytosolic contaminations, respectively. C, Western blotting of total lysate (L), flow-through (F) and elution (E) fractions. CLPX and C1QBP/P32 were used as positive controls for proteins known to interact with Clp(X)P. * represents unspecific antibody binding. D, Scatter plot of proteins co-enrichment in immunoprecipitates of FLAG-CLPP-TRAP and FLAG-CLPP-WT compared with control. Red dots indicate significantly enriched in CLPP-TRAP over CLPP-WT, blue dots proteins significantly enriched with CLPP-TRAP over negative control. The tight binding ATPase subunit CLPX is highlighted in violet. E, Western blotting of POLDIP2 steady state levels in heart lysates, HSC70 was used as control.
