Fig. 7.
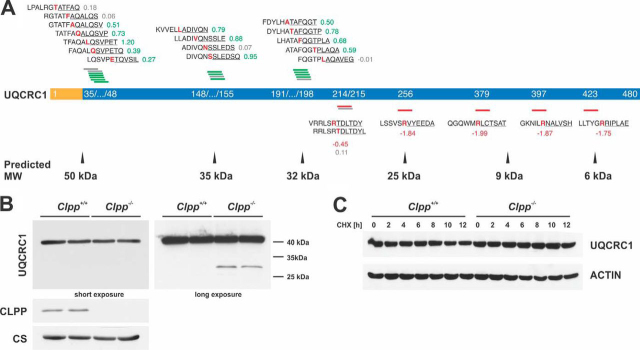
Validation of UQCRC1 as an unexpected ClpXP substrate.A, UQCRC1 N termini abundance. MTS (yellow) and mature protein (blue) are shown with starting position of detected N termini. Above and below the protein, identified N-terminal peptides (underlined) are shown with the associated log2(Clpp−/−/wt) and the preceding sequence. Amino acids at the P1 position are highlighted in red. N termini with increased abundance are depicted in green, with decreased abundance in red and unchanged abundance in gray. Predicted masses of the corresponding proteoforms are indicated below the scheme. B, Western blots of UQCRC1 steady state levels in isolated mitochondria. Citrate synthase (CS) was used as loading control. C, CHX chase experiment of UQCRC1 in MEFs. ACTIN was used as loading control (13).