Figure 3. Pseudotime Analyses Identify Transcription Factor Networks Controlling Human Retinal Cell Fate Specification.
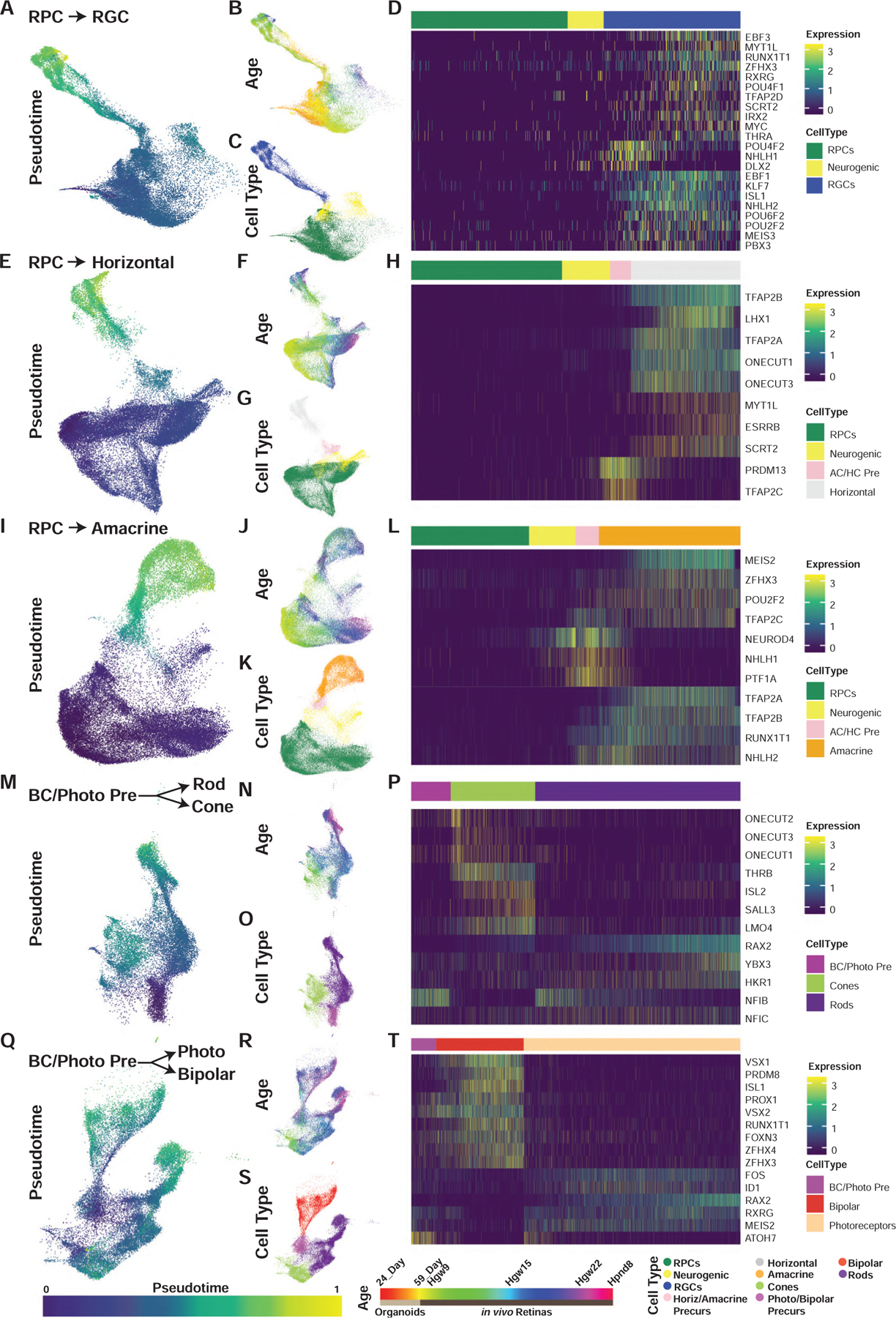
(A–T) UMAP embeddings of cellular inputs for pseudotime analyses of (A–D) retinal ganglion cells, (E–H) horizontal cells, (I–L) amacrine cells, (M–P) rods-cones, and (Q–T) photoreceptor-bipolar cells. UMAP plots are colored by (A, E, I, M, and Q) cellular pseudotime values, (B, F, J, N, and R) age, and (C, G, K, O, and S) cell type. UMAP embeddings shown are subsets of initial dimension reductions. (A–C) contain only RPCs that contribute to the RGC trajectory while (E–T) include all RPCs. (D, H, L, P, and T) heatmap showing relative expression of differentially expressed transcription factors across pseudotime, highlighting transcription factors with enriched expression in endpoint cell types. Abbreviations: Hgw, human gestational weeks; Hpnd, human postnatal day; RPC, retinal progenitor cells; RGC, retinal ganglion cells; Photo, photoreceptors; BC/Photo Pre, bipolar cell/photoreceptor precursors.
