Figure 3. Established transcriptional mechanisms underlying Ikaros family member regulation of target gene expression.
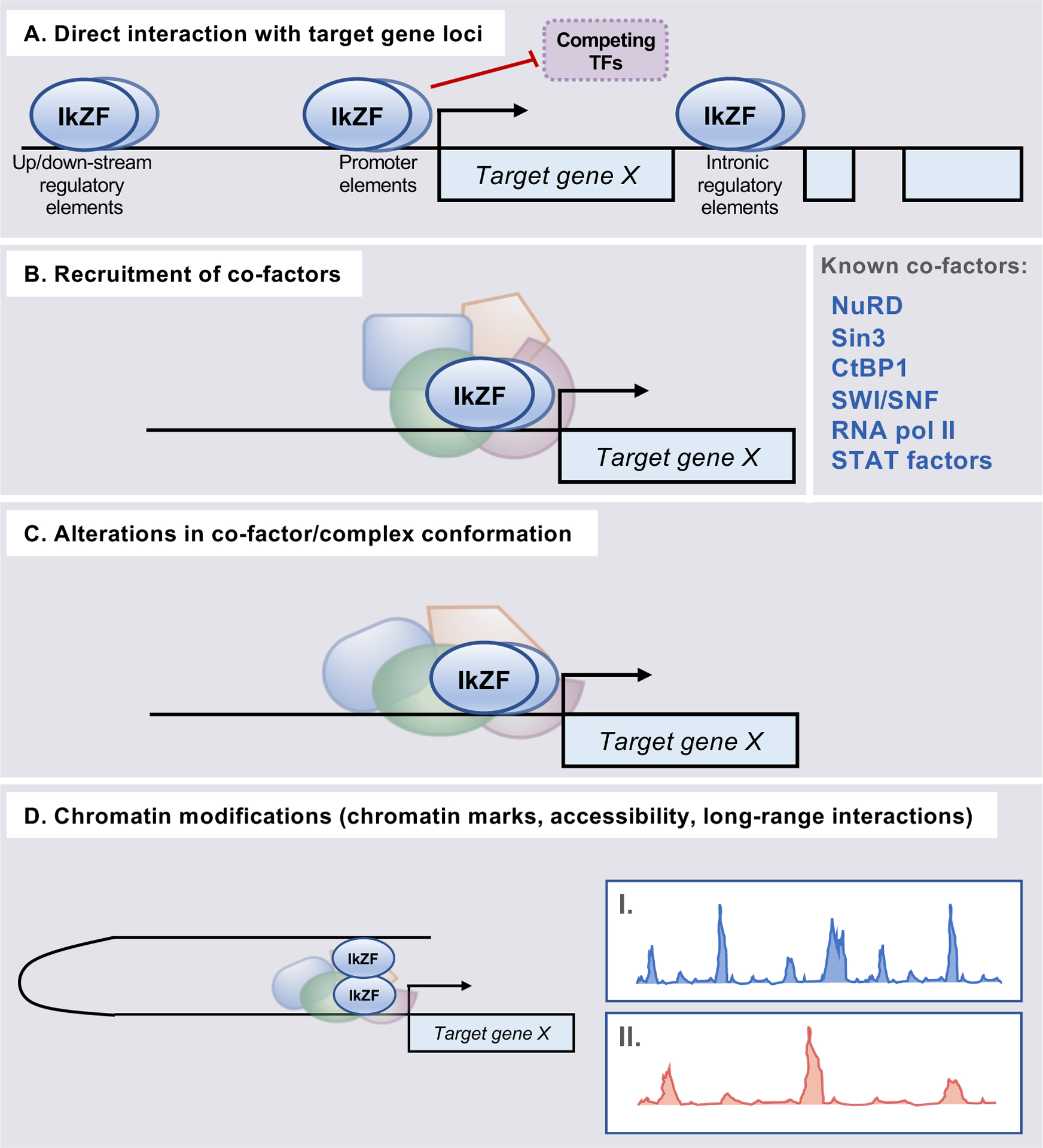
Ikaros family members have long been known to exert transcriptional control through a number of non-mutually exclusive DNA- and chromatin-dependent mechanisms. (A) All family members interact directly with DNA and associate with target gene loci. This allows for locus-specific control via both recruitment of co-factors to target gene regulatory regions, as noted, and by preventing association of competing transcriptional regulators. (B) Ikaros, Aiolos, Helios, and Eos have been found to interact with chromatin remodeling complexes/enzymes and/or additional cofactors to influence the accessibility and expression of target genes. (C) Association of the above co-factors with Ikaros family members has been suggested to result in alterations to co-factor conformation, which alters their function/binding. (D) The interactions described in A-C promote both local and long-range alterations to chromatin structure, including the deposition of chromatin marks (illustrated as example ChIP-seq peaks for chromatin mark enrichment in “I” and “II”). These include changes in regulatory region acetylation and methylation patterns. Ikaros factors have also been implicated in mediating long-range interactions (i.e. chromatin looping) between distal target gene regulatory elements.
