Fig 2. Sequence comparisons of dim2 alleles in Z.
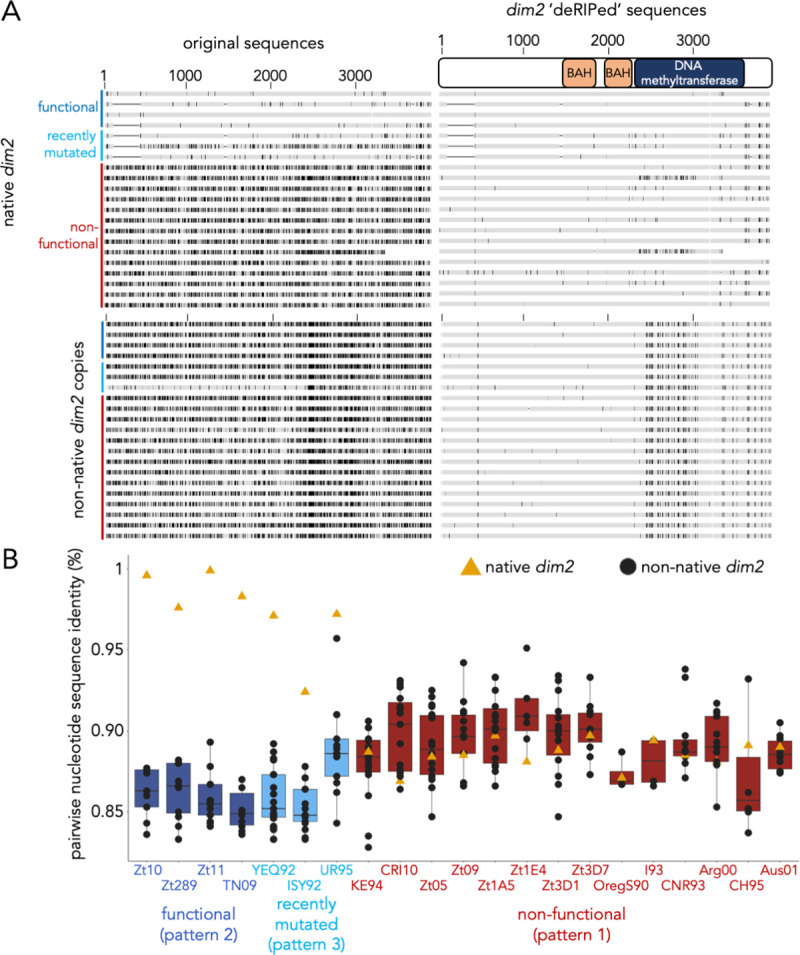
tritici isolates. (A) Mutations of dim2 in original and ‘deRIPed’ alleles identified by alignment to the functional allele of Zt469. Shown are all native alleles (located on chromosome 6) and one representative non-native, additional copy per isolate. Native alleles (functional and non-functional) lack mutations in the DNA methyltransferase domain that are present in all non-native alleles, except for isolates OregS90 and CRI10. (B) Pairwise sequence comparison of the native, intact dim2 allele from Zt469 with all full-length native and additional dim2 sequences (see S2 Table). No additional dim2 copies were detected in the genome of Zt469 (from Aegilops sp.) whereas all other isolates (from Triticum spp.) have additional copies of dim2. Non-native, additional copies (black dots) in the isolates with functional or recently mutated dim2 (except for UR95) showed higher sequence diversity compared to those with inactivated dim2, where alleles at the native locus (yellow triangle) are amongst the more diverged copies. This supports the idea that this is the ancestral, now heavily mutated allele of dim2. Alleles or isolates are arranged in the same order in panels A (top to bottom) and B (left to right).
