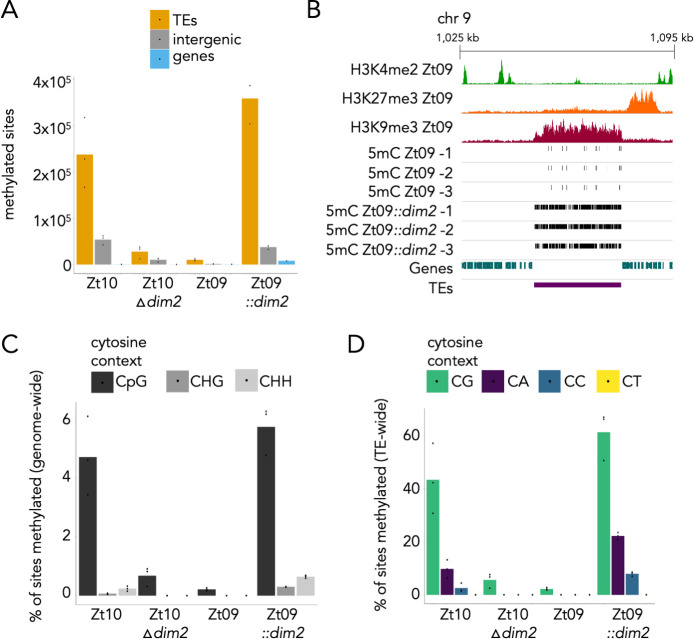
Fig 3. Localization and site preferences of cytosine methylation in Z.
tritici genomes in presence and absence of dim2. (A) Number of 5mC methylated sites detected by whole genome bisulfite sequencing (WGBS) in TEs, intergenic regions, or genes. The vast majority of methylated sites are localized in TEs followed by intergenic regions and TE-derived genes. (B) 5mC co-localizes with H3K9me3 and TEs in Zt09. Shown are ChIP-seq tracks for H3K4me2, H3K27me3 and H3K9me3 [41], and 5mC sites detected in a representative region on chromosome 9 in the genomes of Zt09 and Zt09::dim2 (three replicates). (C) Genome-wide 5mC levels in the different isolates and respective mutants. 5mC levels are higher in the presence of a functional dim2, and dim2 is required for all non-CG methylation. (D) 5mC levels and site preferences in TEs. CG sites are preferred, followed by CA, and CC. 5mC is almost completely absent from CTs.