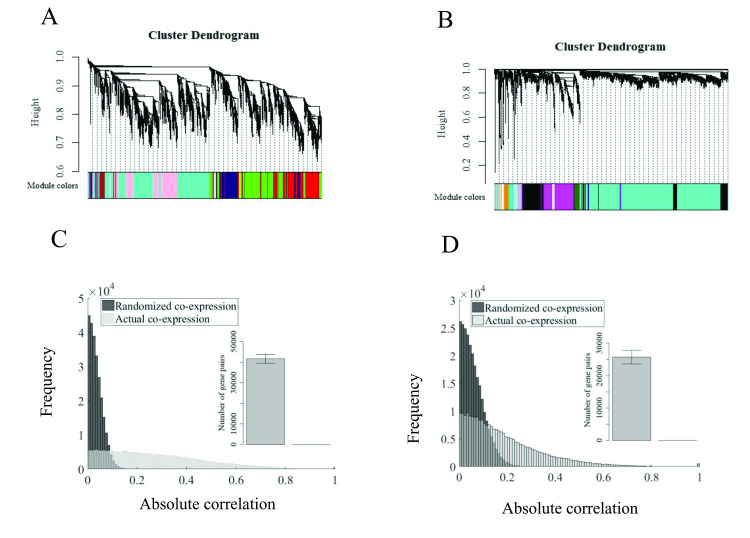
Fig 1. Frequency of co-expression is higher than expected by chance in natural transcriptomes.
A and B) Typical co-expression structure and modular organiza-tion of a random sample of 1000 genes from the human brain transcriptome (Brain-span dataset) and 60 brain tissue samples obtained from the Fantom5 dataset. C) and D) Distribution of absolute correlations of all gene pairs in a random sample of 1000 genes compared with the distribution resulting from random permutations of expression data in the same genes. Inset: Bars show the mean (±SEM) number of highly correlated pairs (/R/>0.5) among 1000 independent samples of 1000 genes each compared with the expected mean number of highly correlated pairs (absolute correlation) resulting from random permutations of the gene expression values in the same samples.