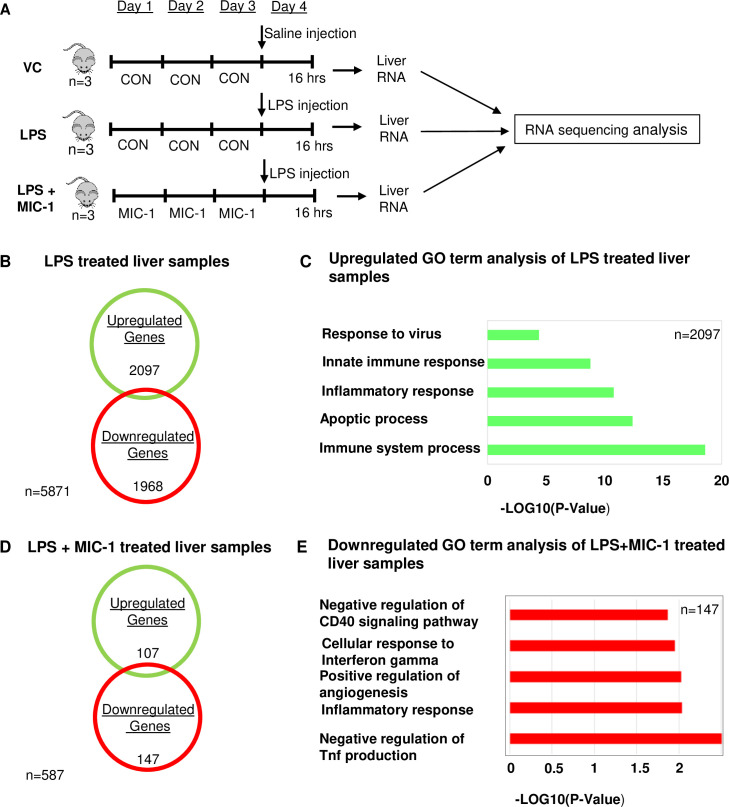
Fig 3. Transcriptome analysis in the liver.
(A) Schematic representation of the course of the experiment. Male C57BL/6 mice (n = 18) were divided into three groups: a. vehicle control (10% DMSO), b. LPS (10 mg/kg), c. LPS (10 mg/kg) + MIC-1 (80 mg/kg). LPS/Saline treatment was given after 3 days of gavaging MIC-1; the liver was collected after 16 hours for RNA sequencing (n = 6). (B) Venn diagram displaying the number of inducible or repressible (P-value < 0.05 and > 1.0 absolute log2-fold change) genes after LPS treatment in the liver. (C) Gene Ontology analysis using DAVID v6.8 functional annotations (biological process) up-regulated by LPS treatment only in liver samples (P-value < 0.05, log2-fold change > 1.0). (D) Venn diagram displaying the number of inducible or repressible (P-value < 0.05 and > 1.0 absolute log2-fold change) genes after LPS+MIC-1 treatment in the liver. (E) Gene Ontology analysis using DAVID (version 6.8) functional annotations (biological process) down-regulated by LPS+MIC-1 treatment in liver samples (P-value < 0.05, log2-fold change < -1.0).