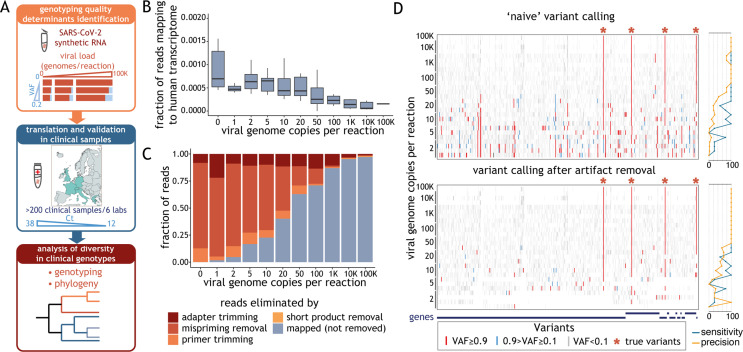
Fig. 1.
Artefact removal is a prerequisite for reliable variant calling. (A) Schematic representation of the study. In experiments using synthetic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA, we varied a number of experimental parameters—including viral load, variant allele fraction (VAF) and sequencing depth—and determined which of these factors critically impact(s) genotyping quality (top box). We validated these metrics using data obtained from clinical samples, whose viral load is reflected by the cycle threshold (Ct) value (middle box). We determined the phylogeny of all clinical samples that met our guidelines (bottom box). (B) Distribution of the fraction of raw reads aligning to human transcriptome (y-axis), obtained with STAR aligner, as a function of the number of synthetic viral genome in the sample (x-axis). The horizontal line in the boxplot indicates the median and the whiskers the 5% and 95% quantile. (C) Average fraction (from at least three replicates) of sequencing reads that mapped to the SARS-CoV-2 genome or were the result of different technical artefacts (y-axis) for samples with varying amounts of synthetic viral genomes (x-axis). (D) Ideogram depicting the location of variants detected in samples with a varying number of synthetic viral genomes (denoted on the left) before (top panel) and after (bottom panel) removal of reads labelled as technical artefacts. Variants with allele fraction <0.1, between 0.1 and 0.9, and >0.9 are shown in grey, blue and red, respectively. Expected SARS-CoV-2 variants present in the control are marked with asterisks. Plots on the right show sensitivity and precision of the variant calls.