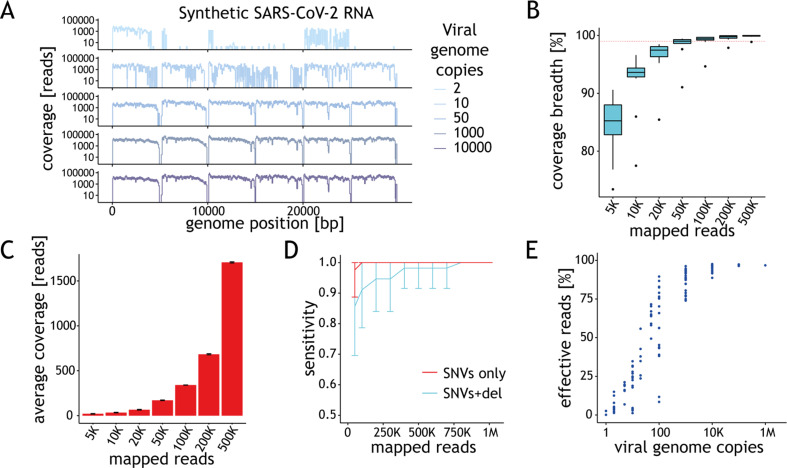
Fig. 2.
Performance of the assay depends on the amount of starting material. (A) Ideograms depicting the genome coverage (y-axis) for representative samples with varying amount of synthetic viral genomes (x-axis). Signal drops every 5 kb are expected due to gaps in the reference material. (B) Distribution of the genome coverage breadth (y-axis) as a function of the number of mapped reads for samples with 10 000 genome copies per reaction (g.c.p.r.). Horizontal dashed line depicts 98% coverage breadth. The horizontal line in the boxplot indicates the median and the whiskers the 5% and 95% quantiles. (C) Average coverage depth across synthetic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome (y-axis) as a function of the number of mapped reads (x-axis) based on data from samples with 10 000 g.c.p.r. (D) Average sensitivity of variant calling for single nucleotide variants (SNVs) (red) or SNVs + 10 bp indel (cyan) in SARS-CoV-2-c1 (y-axis) as a function of the number of mapped reads based on the results obtained for samples with at least 98% genome coverage breadth. Error bars represent standard deviation. (E) Percentage of effective reads (y-axis) shown as a function of the viral load (g.c.p.r.) in the sample. Each point represents the data for one sample.