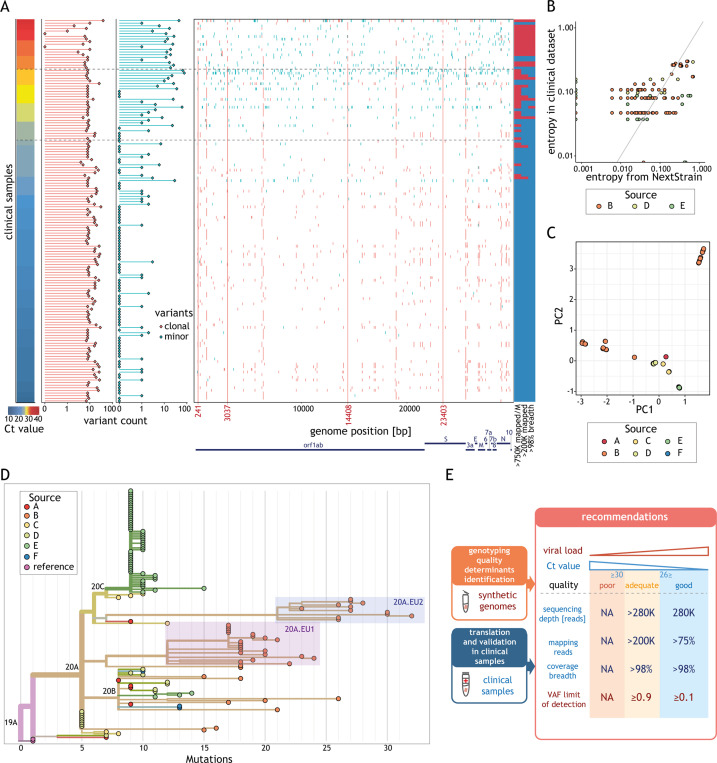
Fig. 5.
Variant frequencies found in the clinical dataset reflect global frequencies. (A) Summary of the variant calling analysis for all unique clinical samples (rows) sorted by the cycle threshold (Ct) value (left). The horizontal dashed lines indicate Ct values of 26 and 30. The numbers of clonal (variant allele fractions, VAF ≥ 0.9, red) and minor (0.1 < VAF < 0.9, cyan) variants for each sample are represented as horizontal bar-plots (middle left). The position of each clonal (red) and minor (cyan) variant is displayed along the genome (middle right). Coordinates marked in red indicate positions of the most prevalent variants. Classification of the samples relative to the different recommendations (listed below each column) (right): blue indicates the recommendation was fulfilled and red that it was not. (B) Relationship between the entropy estimated for all clonal variants in clinical samples (y-axis) and the entropy of the same variants in samples collected in the same country and during the same period according to Nextstrain [30] (x-axis). Only samples with >200 K effective reads and 98% coverage breadth from centres with data for more than 15 samples were considered in this analysis. (C) 2-D principal component analysis results of clonal variants in clinical isolates (points). Points are coloured based on the sample source. (D) Phylogenetic tree of all clinical isolates with >200 K effective reads and 98% coverage breadth criteria. Samples are coloured according to the source. Clades (according to Nextstrain) are indicated. Samples corresponding to subclade 20A.EU.1 and 20A.EU.2 are highlighted by red and blue boxes, respectively. Length of the branches reflects the number of mutations (x-axis). The tree visualization was generated using the Nextstrain platform [30]. (E) Schematic representation of the recommendations for reliable genotyping with amplicon-based approach. We used synthetic viral genomes to determine the minimal viral load and VAF. We validated these recommendations and made them broadly applicable using clinical samples by determining the minimal sequencing depth, fraction of mapped reads and coverage breadth. Samples were classified into three quality categories based on their viral load: good (≥1000 genome copies per reaction (g.c.p.r.)), adequate (uncertain g.c.p.r., Ct values in the range 26–30) and poor (<100 g.c.p.r., typically value Ct > 30).