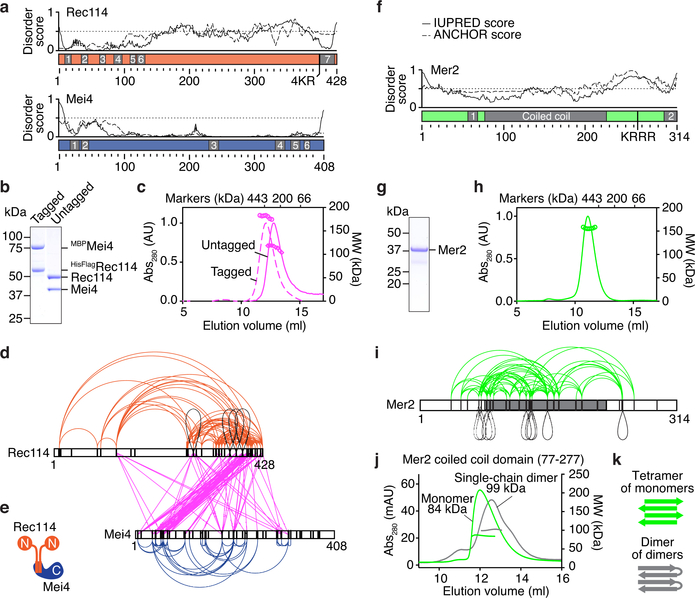
Fig. 1: Purification and subunit arrangement of the S. cerevisiae RMM proteins.
a. Prediction of protein disorder (IUPRED server42). The ANCHOR score predicts the transition from unstructured to structured depending on a binding partner. Previously identified SSMs are highlighted23,26. b. SDS-PAGE of purified tagged and untagged Rec114–Mei4 complexes. 4 μg was loaded. c. SEC-MALS analysis of tagged and untagged Rec114–Mei4. The traces show UV absorbance (left axis), circles are molar mass measurements across the peak (right axis). Elution positions of protein standards are marked. d. XL-MS analysis of Rec114–Mei4 (4812 crosslinked peptides, 258 distinct crosslinked pairs of lysines). Black loops are intermolecular self-links. Black vertical lines indicate lysines. e. Cartoon of the Rec114–Mei4 complex. f. Protein disorder prediction for Mer2. The predicted coiled coil and previously identified SSMs are highlighted23,26. g. SDS-PAGE of purified Mer2. 4 μg was loaded. h. SEC-MALS analysis of Mer2. i. XL-MS analysis of Mer2 (487 crosslinked peptides, 89 distinct crosslinked pairs of lysines). j. SEC-MALS analysis of the coiled coil domain of Mer2 and a single-chain dimer variant of the coiled coil domain. A tetramer of monomers and a dimer of single-chain dimers both have an expected MW of 70 kDa. The difference between the profiles of the monomer and single chain dimer can be explained by reduced degrees of freedom (tension) in the single-chain dimer and heterogeneity. k. Interpretive cartoon of the molecular arrangement of the coiled coil domain of Mer2. For gel source data, see Supplementary Figure 1.