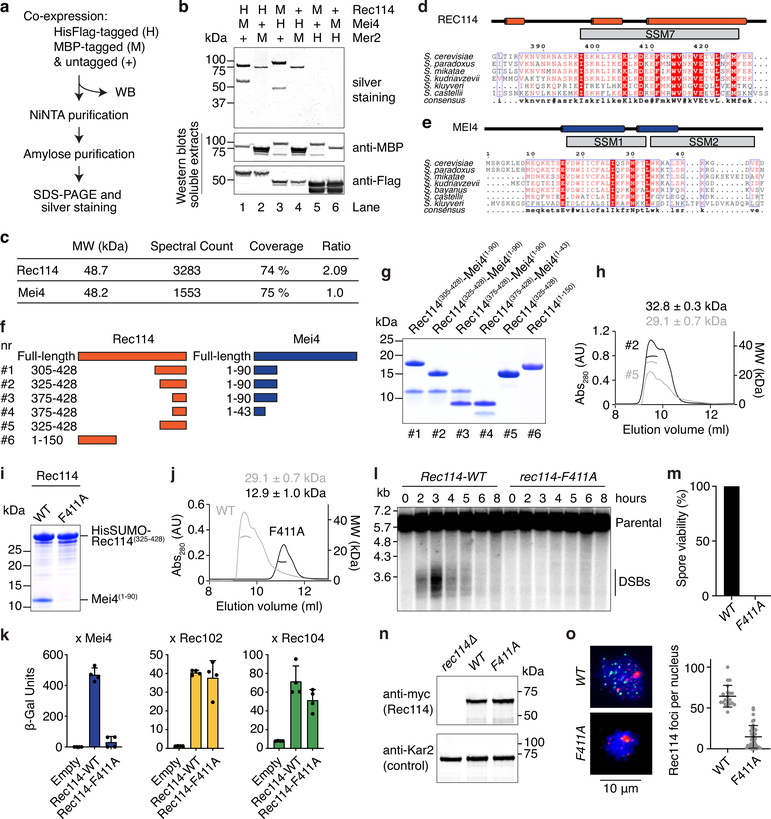
Extended Data Fig. 1: Characterization of the Rec114–Mei4 complex.
a. Strategy for purification of a hypothetical Rec114–Mei4–Mer2 (RMM) complex. Combinations of MBP-tagged and HisFlag-tagged RMM subunits were co-expressed in insect cells. After cell lysis, complexes were purified by sequential affinity chromatography and analyzed by SDS-PAGE. Expression and solubility of the recombinant proteins are verified by western blotting (WB) of cell extracts. b. Analysis of purified complexes. Rec114–Mei4 complexes were apparent (lanes 1 and 3), but no Mer2 was co-purified. Lanes 2 and 4 show some enrichment of MBP-Mer2, but no co-purification of Rec114–Mei4. The presence of MBP-Mer2 in lanes 2 and 4 of the silver-stained gel may be due to background binding of MBP-Mer2 to the NiNTA resin (potentially via adsorption of DNA to the resin), or to low-affinity interactions to immobilized His-tagged Rec114–Mei4 complexes. Either way, none of the combinations tested yielded stoichiometric complexes of all three RMM subunits. Western blot controls of cell extracts showed that the tagged RMM proteins were expressed and soluble. c. Mass spectrometry analysis of Rec114–Mei4 complexes. Purified Rec114-Mei4 complexes were treated with trypsin and analyzed by LC-MS/MS. The ratio of spectral counts between Rec114 and Mei4 provides additional evidence supporting the 2:1 stoichiometry of the complex. d, e. Alignments and predicted secondary structures of the C-terminus of Rec114 (d) and the N-terminus of Mei4 (e). The positions of the conserved SSMs are indicated. f. Cartoon of the Rec114–Mei4 truncations analyzed. g. Purification of Rec114–Mei4 truncations. Proteins were expressed in E. coli and purified on NiNTA resin using a HisSUMO tag fused to the N-terminus of the Rec114 fragment. After removal of the tag by treatment with the SUMO protease Ulp1, complexes were further purified by gel filtration. A Coomassie-stained SDS-PAGE analysis of purified complexes is shown. 5 μg was loaded for each sample. Polypeptides containing Rec114(375−428) and Mei4(1−43) retained the ability to interact (combination #4). h. SEC-MALS analysis of Rec114–Mei4 truncations. The data are consistent with expectation for truncations that contain two Rec114 subunits and one Mei4 subunit. The C-terminus of Rec114 alone forms a dimer. i. Wild type and F411A-containing variants of HisSUMORec114(325−428) were co-expressed with Mei4(1−90) and purified by chromatography on NiNTA resin. The absence of the Mei4 fragment with Rec114-F411A shows that the mutation abolishes the interaction with Mei4. j. SEC-MALS analysis of untagged wild-type (WT, reproduced from panel H to aid comparison) and F411A Rec114(325−428) show that the mutation affects Rec114 dimerization. k. Y2H analysis of the interaction of Gal4BD-Rec114 (WT and F411A) with LexA-Mei4, LexA-Rec102, or LexA-Rec104 (mean and SD from four replicates). β-Gal units are quantified based on hydrolysis of ONPG. The F411A mutation abolishes the interaction of Rec114 with Mei4, but not with Rec102 and Rec104. l. Southern blot analysis of meiotic DSB formation at the CCT6 hotspot, showing that rec114-F411A is defective in meiotic DSB formation. m. Spore viability of rec114-F411A mutant (n = 40). n. Western-blot analyses of meiotic protein extracts from myc-tagged REC114-WT and F411A strains. The F411A mutation does not compromise the expression of Rec114. o. Immunofluorescence microscopy analysis of meiotic chromosome spreads with wild-type and F411A myc-tagged Rec114. Green, anti-myc; red, synaptonemal complex component Zip1; blue, DNA. Quantification of the number of Rec114 foci per leptotene or early zygotene cell is plotted; error bars show mean ± SD (n = 20 and 38 cells for WT and F411A, respectively). The F411A mutation abolishes the formation of chromatin-associated Rec114 foci.