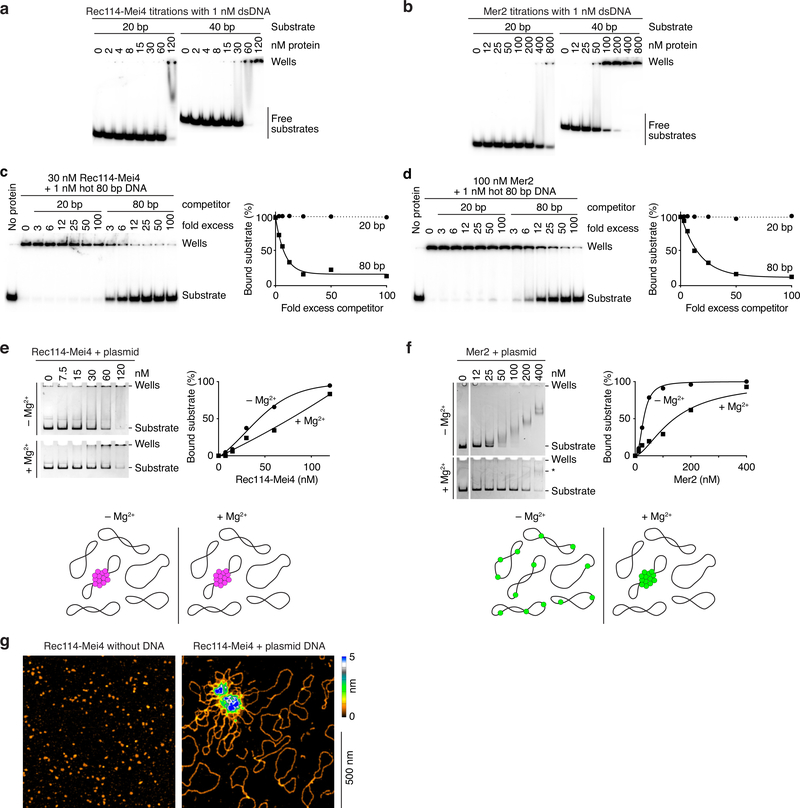
Extended Data Fig. 2: DNA-binding properties of Rec114–Mei4 and Mer2 complexes.
a, b. Gel shift analysis of Rec114–Mei4 (a) or Mer2 (b) binding to 20- or 40- bp DNA substrates. Quantification is in Fig. 2b. c, d. Competition assay of Rec114–Mei4 (c) or Mer2 (d) binding to 80 bp radiolabeled DNA (1 nM) in the presence of 20- or 80 bp cold competitor. Fold excess is in nucleotides. Lines are one-phase decay fits. e, f. Binding to plasmid DNA analyzed by native agarose gel electrophoresis. Rec114–Mei4 (e) and Mer2 (f) were titrated with 2 nM plasmid DNA (pUC19) in the presence or absence of 5 mM MgCl2. Rec114–Mei4 complexes bound with roughly similar affinity independently of the presence of Mg2+ (apparent KD ≈ 50–80 nM). Note that the apparent affinity is significantly lower than suggested by the gel shift analyses with radiolabeled substrates presented in panel a and Fig. 2a, b (see apparent affinities in Fig. 2 legend). We interpret that this difference is because the proteins coalesce on a small fraction of the plasmid molecules, as illustrated in the cartoon below. Indeed, bound plasmids remained trapped in the wells, which is consistent with cooperative assembly of large nucleoprotein structures. Because each plasmid substrate provides many more binding sites than the short oligonucleotide substrates in panel a and Fig. 2a, a higher concentration of protein is required to reach complete binding of all of the plasmid molecules. In contrast to Rec114–Mei4, Mer2 showed efficient binding in the absence of Mg2+ in this assay (KD = 30 ± 2 nM) but binding appeared to be considerably inhibited in the presence of Mg2+ (KD ≈ 150 nM), as indicated by the persistence of unbound substrate at high protein concentrations. However, while the electrophoretic mobility of Mer2-bound plasmids decreased steadily as the concentration of Mer2 increased in the absence of Mg2+, no such steady progression was observed when Mg2+ was included. Instead, a minority of bound substrates shifted to a low-mobility species (labeled * in panel f, bottom), indicating that they were occupied by multiple Mer2 complexes. We interpret that, rather than inhibiting DNA binding, Mg2+ promotes cooperativity, in agreement with the fluorescence microscopy analysis (Extended Data Fig. 3b). The difference in migration distance of the plasmid between the +/− Mg2+ gels is due to the presence of Mg2+ in the electrophoresis buffer. g. AFM imaging of 12 nM Rec114–Mei4 in the absence (left) or in the presence (right) of 1 nM plasmid DNA (pUC19).