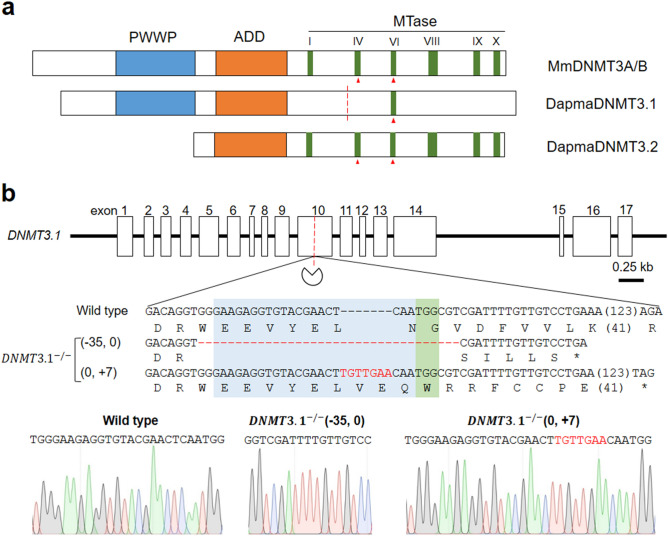
Figure 1.
CRISPR/Cas-mediated mutagenesis of DNMT3.1 (a) Schematic diagrams of domain structures of DapmaDNMT3.1 and DNM3A/B of mouse (Mm = Mus musculus). Red triangles indicate the active sites of catalysis. The red dashed line indicates a targeted disrupting site by CRISPR/Cas system (pie icon). (b) Genomic sequence of the DapmaDNMT3.1 mutant around the gRNA-targeted site. The exons and introns of the DapmaDNMT3.1 (DNMT3.1) were illustrated in white boxes and black lines, respectively. In the alignment, the top line represents the wild type DNMT3.1 nucleotide sequence, and the deduced amino acid sequence is shown under the DNA sequence. Subsequent lines show sequences of mutant alleles followed by deduced amino acid sequences. The asterisk represents a premature STOP codon. The length (base pairs) of an abbreviated sequence and amino acid sequence are shown in parentheses among the sequences. The length (base pairs) of each indel mutation is written in the left of each sequence (−, deletions; + , insertion). The gRNA recognition and protospacer adjacent motif (PAM) sequences are in blue and green boxes, respectively. Red hyphens and letters indicate the identified mutations. Sanger Sequencing chromatograms of DNA from wild type and the biallelic mutant of DNMT3.1 were shown below the alignment.