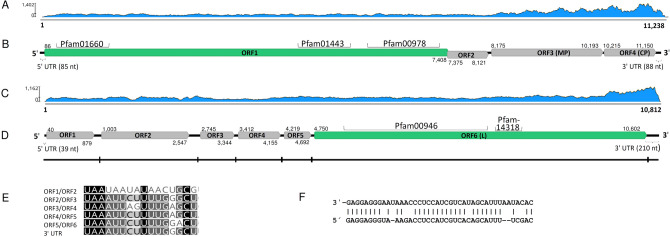
Figure 1.
Genome organization of the Armillaria borealis mycovirgavirus 1 (AbMV1) and Armillaria mellea negative strand RNA virus 1 (AmNSRV1). (A) Coverage of raw reads mapped against the AbMV1 genome (without poly(A) tail). (B) Schematic presentation of AbMV1 genome with UTRs, predicted ORFs and conserved motifs: pfam00978 (RdRP domain, e-value 2.07e−72), pfam01660, (methyltransferase, e-value 1.92e−20) and pfam01443 (helicase, e-value 1.02e−24). Predicted translation initiation and termination sites are marked above or below each ORF. (C) Coverage of raw reads mapped against the AmNSRV1 genome. (D) Schematic presentation of AmNSRV1 genome with UTRs, predicted ORFs and conserved motifs: pfam00946 (Mononegavirales RdRP, e-value 1.10e−95) and pfam14318 (Mononegavirales mRNA-capping region V, e-value 4.82e−16). Predicted translation initiation and termination sites are marked above and below each ORF, respectively. The small vertical lines in the virus genomic segment show the location of the putative gene junction sequences. (E) Consensus sequences (uracil-rich tracts) of putative gene junction regions between the predicted ORFs in 3′ > 5′ orientation (genomic RNA). (F) Complementarity in the 5′- and 3′-terminal sequences in the genomic RNA strand.