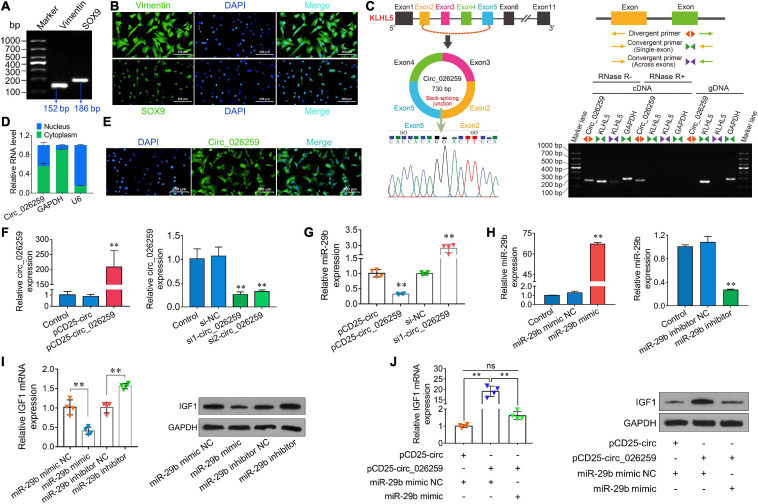
FIGURE 7.
Characterization and expression analysis of circ_026259 and effects of circ_026259 on oar-miR-29b and IGF1 expression in sheep Sertoli cells. (A) RT-PCR and (B) immunofluorescence assays of marker genes (Vimentin and SOX9) were performed to identify the primary Tibetan sheep Sertoli cells. (C) The structure characteristics of circ_026259. (Left upper panel) Schematic illustration showing circularization of circ_026259 originated from exon 2 to exon 5 of the ovine KLHL5 gene. (Right panel) Divergent primers amplified circ_026259 in cDNA in Sertoli cells with or without RNase R, but not genomic DNA (gDNA). (Left lower panel) Sanger sequencing confirmed the back-splicing junction of circ_026259. (D) qPCR detection and (E) FISH localization of circ_026259 in Sertoli cells. (F) Examination of circ_026259 overexpression (left panel) and knockdown (right panel) efficiency by qPCR. (G) Effects of overexpression and knockdown of circ_026259 on expression of oar-miR-29b. (H) Examination of oar-miR-29b overexpression (left panel) and knockdown (right panel) efficiency by qPCR. (I) Effects of overexpression and knockdown of oar-miR-29b on IGF1 mRNA and protein expression. (Left panel) IGF1 mRNA levels relative to GAPDH. (Right panel) the representative images showing IGF1 protein expression levels obtained using Western blot. (J) Circ_026259 overexpression can rescue the suppressive effects of oar-miR-29b on IGF1 mRNA and protein expression in Sertoli cells. (Left panel) the relative mRNA levels of IGF1. (Right panel) the representative Western blot images showing IGF1 protein expression. ∗∗ denotes P < 0.01.