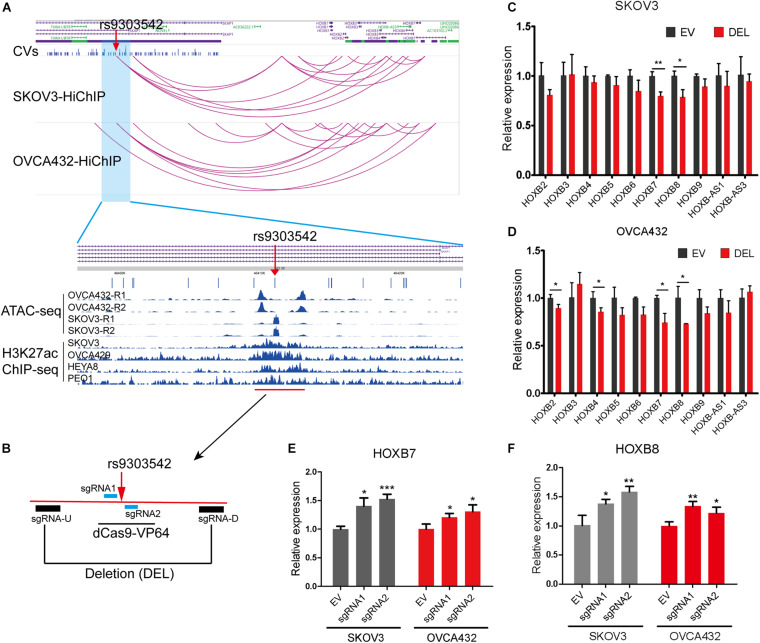
FIGURE 2.
Validation of regulatory relations between rs9303542 enhancer region and HOXB genes. (A) Interaction profiles of rs9303542 and HOXB genes at 17q21.32 and ATAC-seq and H3K27ac-ChIP signal enrichment at the rs9303542 region. (B) A schematic representation elucidating the design for CRISPR-Cas9 deletion and dCas9-VP64 activation experiments. sgRNA-U and sgRNA-D were cloned in px459v2 respectively and then cotransfected into the indicated cells to delete the 2000bp rs9303542 enhancer region. The sgRNA1/2 were separately cloned into MS2-gRNA-hU6 expression vector and then transfected into dCas9-VP64 stable expression SKOV3 and OVCA432 cells. (C,D) qPCR was used to detect the expression of HOXB genes between rs9303542 deleted (DEL) and vector control (EV) cells in SKOV3 (C) and OVCA432 (D). The expression of HOXB1, HOXB-AS2, and HOXB-AS4 was too low to detect in both cell lines. (E,F) qPCR was used to compare the expression of HOXB7 (E) and HOXB8 (F) after sgRNA1/2 transfected (sgRNA1, sgRNA2) and empty vector transfected (EV) cells with dCas9-VP64 stable expression. Error bars, SD. ns: not significant, *p < 0.05, **p < 0.01, ***p < 0.001 as determined by an unpaired, two-tailed Student’s t-test.