Abstract
Rationale: Birth cohort studies have identified several temporal patterns of wheezing, only some of which are associated with asthma. Whether 17q12-21 genetic variants, which are closely associated with asthma, are also associated with childhood wheezing phenotypes remains poorly explored.
Objectives: To determine whether wheezing phenotypes, defined by latent class analysis (LCA), are associated with nine 17q12-21 SNPs and if so, whether these relationships differ by race/ancestry.
Methods: Data from seven U.S. birth cohorts (n = 3,786) from the CREW (Children’s Respiratory Research and Environment Workgroup) were harmonized to represent whether subjects wheezed in each year of life from birth until age 11 years. LCA was then performed to identify wheeze phenotypes. Genetic associations between SNPs and wheeze phenotypes were assessed separately in European American (EA) (n = 1,308) and, for the first time, in African American (AA) (n = 620) children.
Measurements and Main Results: The LCA best supported four latent classes of wheeze: infrequent, transient, late-onset, and persistent. Odds of belonging to any of the three wheezing classes (vs. infrequent) increased with the risk alleles for multiple SNPs in EA children. Only one SNP, rs2305480, showed increased odds of belonging to any wheezing class in both AA and EA children.
Conclusions: These results indicate that 17q12-21 is a “wheezing locus,” and this association may reflect an early life susceptibility to respiratory viruses common to all wheezing children. Which children will have their symptoms remit or reoccur during childhood may be independent of the influence of rs2305480.
Keywords: latent class analysis, genetics, 17q12-21, wheeze, asthma
At a Glance Commentary
Scientific Knowledge on the Subject
Previous studies of childhood wheezing have identified multiple temporal wheeze phenotypes. Although multiple studies have found associations between 17q12-21 SNPs and childhood asthma, associations with different wheeze patterns are understudied.
What This Study Adds to the Field
In this study, we first derive wheeze phenotypes using latent class analysis and then test for associations with 17q12-21 SNPs. We find associations between 17q SNPs and all three wheezing phenotypes. By leveraging the difference in linkage disequilibrium between European American and African American children, we identify the GSDMB SNP rs2305480 as the likely driver of these associations.
Asthma and asthma-like symptoms are a frequent cause of morbidity and healthcare costs during childhood (1). The most common clinical expression of the disease in early life is episodic wheezing, usually as a consequence of viral respiratory illnesses. However, wheezing patterns are highly heterogeneous, and although many children wheeze during the first few years of life, in the majority, these symptoms will remit. Furthermore, a subset of children may not wheeze in the first few years of life but start wheezing or develop asthma later. Recently, a number of studies have used latent class analysis (LCA) to derive data-driven wheeze phenotypes from longitudinally collected wheeze data with comparable results (2–4). Identifying the origins of these patterns may provide insight into asthma pathogenesis as well as provide an opportunity to identify early on those children in greatest need of intervention.
Genome-wide association studies performed over the last decade have found a number of associations between SNPs and childhood onset asthma, with the most significant and widely replicated locus being on chromosome 17q12-21 (5). Only one study (6), however, has examined associations between LCA-derived wheezing phenotypes and 17q12-21 SNPs. Furthermore, most genetic studies have included predominantly participants of European ancestry, in which linkage disequilibrium (LD) at this locus extends for over 200 Kb, hampering efforts to identify the specific variants responsible for the strong asthma associations in this region.
A multiancestry study suggested that this locus may harbor several independent asthma-related variants (7), but whether these putative loci underlie different clinical expressions of the disease remains undetermined. Ober and colleagues (8) recently leveraged the breakdown in LD in African American (AA) children to fine-map the region by looking at associations between childhood onset asthma (before age 6) and 17q12-21 SNPs in both AA and European American (EA) children. However, no study has yet assessed associations between 17q12-21 SNPs and wheezing phenotypes in AA children.
The CREW (Children’s Respiratory Research and Environment Workgroup) (9) is a consortium of birth cohort investigative teams previously funded to study asthma and allergy that are now part of the NIH’s Environmental Influences on Child Health Outcomes initiative. This consortium offered a unique opportunity to characterize wheezing phenotypes and determine associations between longitudinal wheeze patterns and chromosome 17q12-21 SNPs in both AA and EA children. Here, we focus on wheezing patterns in early life and childhood. Using data from seven CREW cohorts, we first harmonized the outcome of reported wheeze in 3,786 children from birth to age 11. After using LCA to derive wheeze phenotypes, we determined associations between the phenotypes and nine SNPs spanning the 17q12-21 locus, which were chosen based on associations with childhood asthma in earlier studies. We hypothesized that some patterns of wheezing, such as a persistent wheeze, in the first decade of life would be associated with genetic variants on chromosome 17q12-21 and that the breakdown of LD in the AA children would allow us to better distinguish the key SNPs.
Methods
Data Harmonization
Data from seven CREW birth cohorts were included in this analysis. Information on each cohort is provided in Table E1 in the online supplement. The CREW cohorts collected information from their study participants using various questions administered at different ages. Therefore, it was necessary to harmonize the data across study participants in the cohorts. This process required addressing different lengths of follow up as well as systematic missingness owing to study protocols in which participants were not contacted at specific ages. More detail on the harmonization process is provided in the supplemental Methods. For wheeze, all cohorts regularly asked questions about whether the subject wheezed over some period of time (e.g., the last 12 mo). Using those responses, we created binary variables indicating whether a subject wheezed in each year from birth until age 11. Study participants with fewer than three responses to wheeze questions through age 11 were excluded from the main analysis. Harmonized variables were also created for race/ancestry, exposure to a dog or cat in the first year of life, and exposure to smoke during the prenatal period. An asthma variable was harmonized from parental reports of doctor-diagnosed asthma up to age 11. Total IgE values, available for 2,010 subjects from six of the seven cohorts, were also harmonized.
Genotyping and Genotype Quality Control
Nine SNPs, rs2941504, rs2517955, rs12936231, rs2305840, rs7216389, rs4065275, rs8076131, rs8069202, and rs3859192, were selected from the 17q12-21 locus based on previously reported associations and LD patterns. The supplemental Methods and Table E2 provide additional information on specific SNPs used in this study. DNA (500 ng at concentrations of 5 ng/μl) was shipped from the coordinating center of each of the cohorts to the University of Chicago for genotyping using TaqMan assays (Applied Biosystems) and standard quality control checks. Call rates were >95% for all SNPs.
Statistical Analyses
For each subject with at least three wheeze data points (n = 3,786), we constructed a wheeze “pattern” based on the binary variables indicating whether or not the child wheezed in each year. We then performed an LCA on those data using the poLCA R package (10) to derive longitudinal “wheeze phenotypes” represented by the different latent classes. Because of substantial uncertainty in latent class membership (discussed further in the supplemental Methods and Results), we did not classify subjects to their highest posterior probability class but instead employed a novel compositional data analysis approach (11, 12) using the full vectors of posterior probabilities, allowing us to include class uncertainty in the analysis. To assess the robustness of the LCA results, we repeated the LCA multiple times with various subsets, including running the LCA separately for AA and EA children. See the supplemental Methods for more detail.
We tested for associations between latent class membership and our nine a priori selected SNPs using a compositional data analysis multiple regression framework. An additive genotype model was assumed for each SNP. Additional details of the analyses and a discussion of multiple comparisons are available in the supplemental Methods.
Results
Characteristics of the Study Population
A total of n = 3,786 children had at least three harmonized wheeze data points and were included in the wheezing LCA. Basic demographic information for the seven cohorts is presented in Table 1; additional information on the distribution of wheeze data by cohort and missing values are presented in Figures E1 and E2 and Table E5. Genotypic data were available for 72% of participants. Only two racial/ancestry groups had sufficient sample sizes for genetic association studies: AA (n = 620) and EA (n = 1,308). No siblings were included in the genetic association studies.
Table 1.
Cohort Demographics
| Latent Class Analysis |
Genotype Analysis |
|||
|---|---|---|---|---|
| n | % | n | % | |
| Total subjects | 3,786 | — | 1,928 | — |
| Sex | ||||
| F | 1,865 | 49 | 917 | 48 |
| M | 1,921 | 51 | 1,011 | 52 |
| Race/ancestry | ||||
| African American | 772 | 20 | 620 | 32 |
| European American | 1,962 | 52 | 1,308 | 68 |
| Mexican American | 288 | 8 | — | — |
| Puerto Rican/Dominican | 450 | 12 | — | — |
| Other | 314 | 8 | — | — |
| Prenatal ETS exposure | ||||
| None | 3,323 | 91 | 1,725 | 89 |
| 1–10 cigarettes/d | 237 | 7 | 156 | 8 |
| >10 cigarettes/d | 78 | 2 | 47 | 2 |
| Infant dog exposure | 950 | 26 | 588 | 30 |
| Infant cat exposure | 659 | 18 | 448 | 23 |
Definition of abbreviation: ETS = environmental tobacco smoke.
The latent class analysis (left) used all subjects with at least three wheeze data points, whereas the genotype association analyses focused on African American and European American children because of small sample sizes in other groups. Some respondents did not have data for whether the mother smoked during pregnancy (prenatal ETS exposure).
Wheezing Patterns
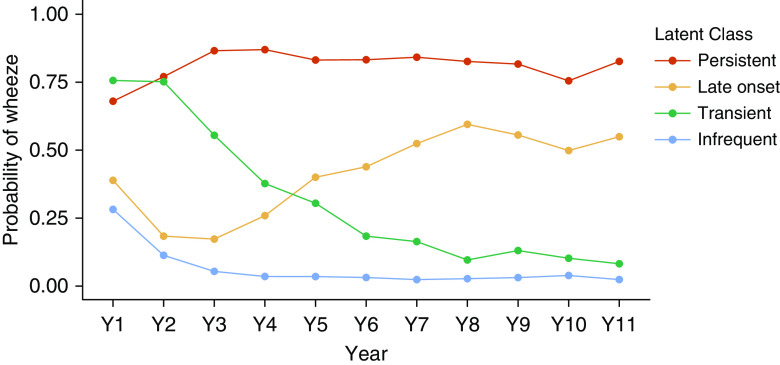
Overall, 52% of children wheezed at least once in the first 3 years of life, and 62% wheezed in the first decade of life. Using the Bayesian information criterion (BIC), we identified four latent classes that differ in the probability of wheezing during each year of life (Figure 1), labeled as infrequent, transient, late-onset, and persistent wheeze. Similar patterns have been identified for several of the seven cohorts (13–16). Despite substantial missing data because of cohort heterogeneity, repeated analyses with differing amounts of missing data consistently identified four classes with very similar structure, with the exception of the LCA of complete cases (n = 601), which had a lower BIC for three classes. Separate analyses of EA and AA children also found four classes with a similar structure. Excluding cohorts, one at a time and rerunning the analysis resulted in classes similar to those defined for the larger group. For all relevant analyses, we chose the infrequent class as the reference category.
Figure 1.
Results of the latent class analysis of wheeze data from CREW (Children’s Respiratory Research and Environment Workgroup) children (n = 3,786). The latent class analysis was performed on binary strings representing whether a child wheezed in each year of life from birth to age 11 years. Four classes were chosen using the Bayesian information criterion. The probability of wheezing in each year of life is shown for each class using the following colors: red = persistent, yellow = late onset, green = transient, and blue = infrequent.
Classifying subjects to their highest posterior probability class, we found 62% were classified as infrequent, 17% as transient, 10% as late-onset, and 11% as persistent. However, many individuals were not classified with a high probability to any single class, and one-third of subjects had a maximum posterior probability less than 0.80 (Figure E3), indicating that many observed wheeze patterns were compatible with multiple classes. Subjects with more missing data tended to have lower maximum posterior probabilities (Figure E4), but some subjects with complete or nearly complete data also had low maximum posterior probabilities, indicating an ambiguous wheeze trajectory. For example, a subject who wheezed only in Years 2 and 6 might be assigned a 0.6 probability of being from the infrequent class and a 0.4 probability of being from the transient class, indicating that the wheeze pattern identified is compatible with both classes. To incorporate that class membership uncertainty into the genetic association analysis, we did not classify individuals to their highest probability class but instead used the vector of four latent class membership posterior probabilities directly in the statistical models.
As expected, the odds of latent class membership were associated with sex, race/ancestry, smoke exposure, and total IgE. Details of those results are presented in the online supplement. Compared with EA children, AA children had a greater probability of being assigned to the persistent class (16% vs. 10%; P = 0.0001) but did not differ significantly from EA in assignment to the other wheeze classes (18% vs. 18%, P = 0.33 for transient; 13% vs. 10%, P = 0.11 for late-onset).
Relationship of 17q12-21 Genotypes to Wheezing Phenotypes
Most 17q SNPs were associated with higher odds ratios (ORs) for all three wheezing phenotypes in subjects of both African and European ancestry (Figure 2 and Table E6). In general, for each SNP, the ORs were greatest for persistent wheeze and lowest for transient wheeze, but for some SNPs (e.g., rs7216389), the ORs for transient wheeze exceeded late-onset wheeze.
Figure 2.
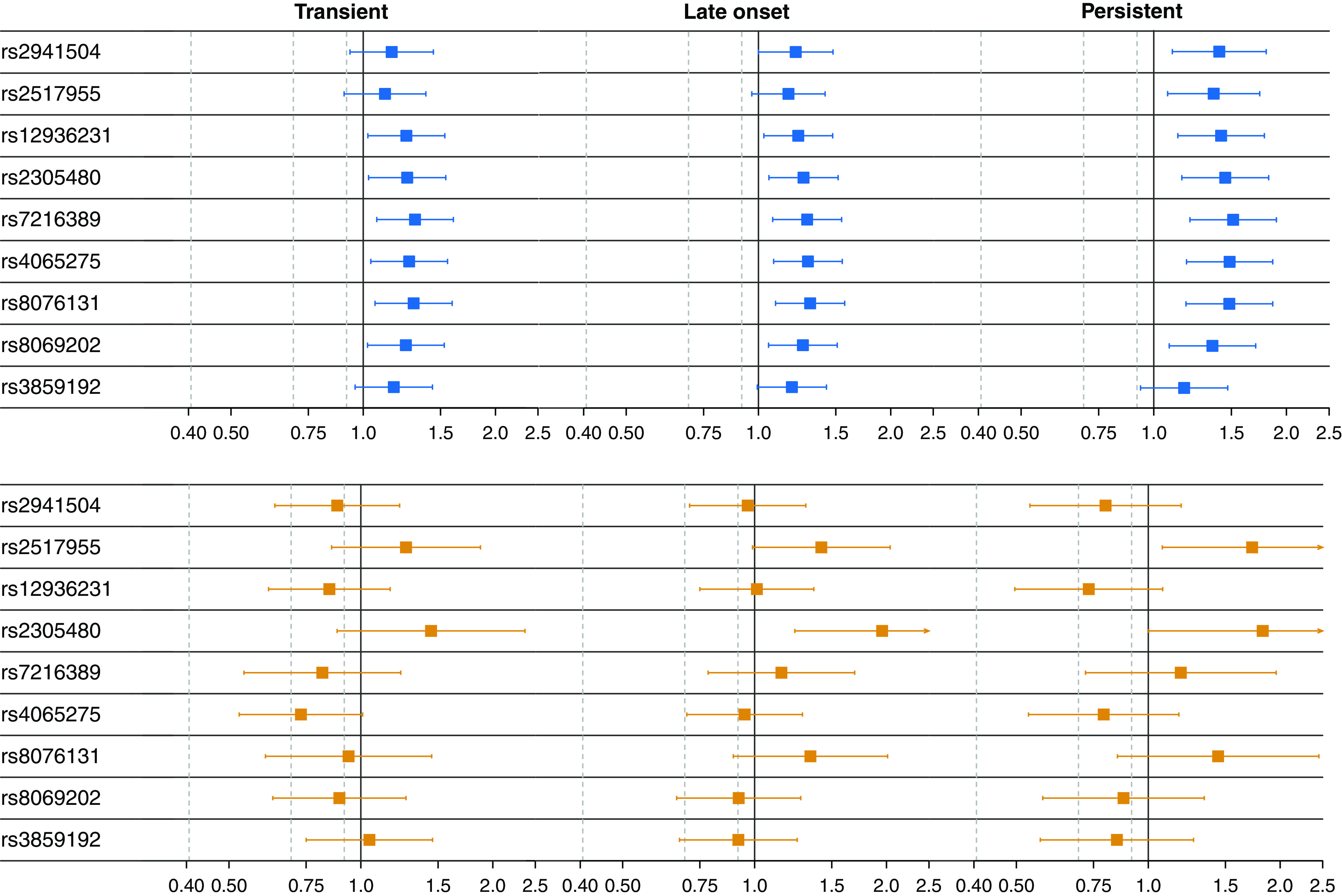
Associations between wheezing phenotypes and SNPs in the 17q12-21 region for European American (EA) (n = 1,308) and African American (AA) (n = 620) children. The panels show the estimated odds ratios on a log scale for being in the three latent wheezing classes (persistent, late onset, or transient) versus the infrequent class by SNP, separately for EA (top, blue) and AA (bottom, orange). Here, an odds ratio greater than one indicates that having the risk allele (see Table E2) was associated with higher odds of belonging to the latent wheezing class relative to the infrequent class. The bars illustrate the 95% confidence intervals. A small arrow at the end of a confidence interval indicates that the interval continues to the right but was cut off for visualization.
For the EAs, the odds of being in any of the wheezing classes was higher for all SNPs except rs3859192, although in some cases, the confidence intervals included one (i.e., equal odds) for either the late-onset or transient classes. The pattern was starkly different for the AAs, in whom many SNPs did not show associations with any wheezing phenotype. For both AA and EA, however, the strongest signals were with the same two SNPs, rs2517955 in PGAP3 and rs2305480 in GSDMB, which had the largest ORs for all three latent class comparisons. These SNPs are in stronger LD (Figure 3) in African Americans (r2 = 0.40) than in Europeans (r2 = 0.30). The point estimates for the ORs for these two SNPs were larger for AA than EA, despite the confidence intervals crossing 1.0 in AA, likely because of the smaller sample size of AA subjects. Two other SNPs, rs7216389 and rs8076131, showed inconsistent signals across the classes in AA subjects.
Figure 3.
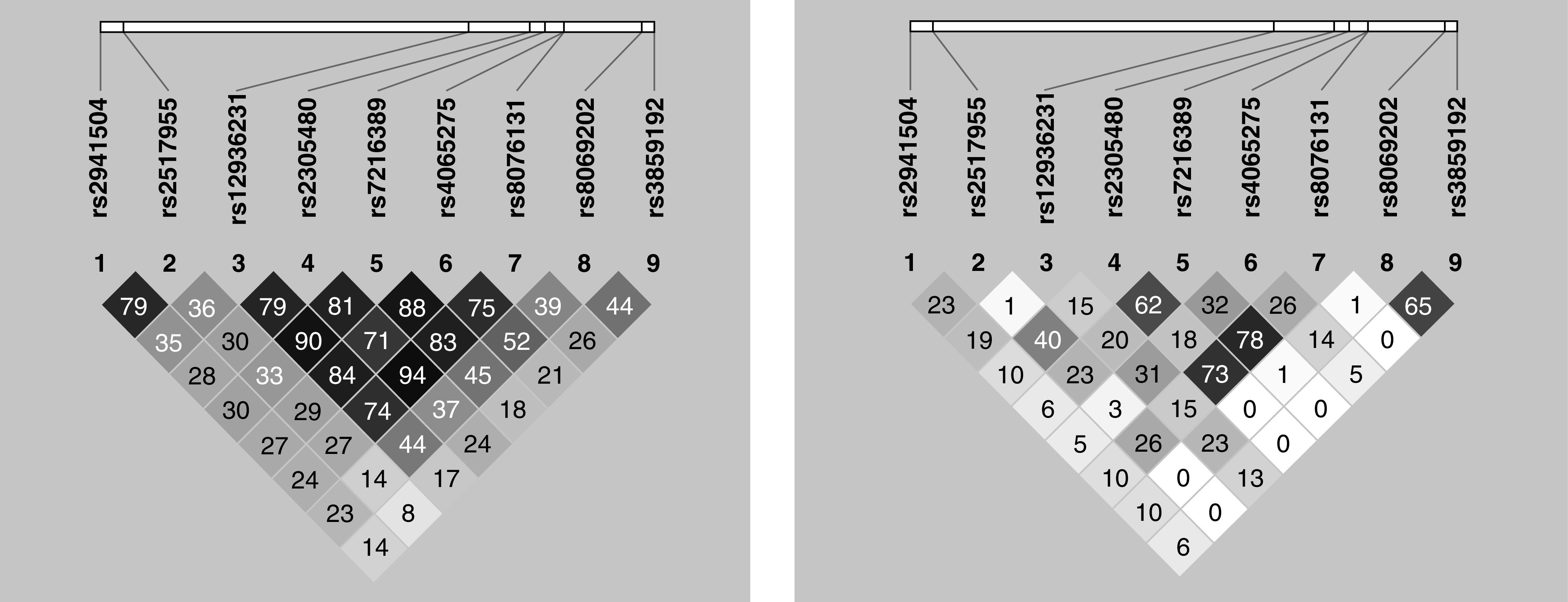
Patterns of linkage disequilibrium for the nine SNPs in both European American (left) and African American children (right). The r2 values for pairs of SNPs are shown within the diamond shapes for all values <1.0.
Table E6 also shows the results of two adjustments for multiple testing, including the conservative Bonferroni method and one based on the effective number of SNPs (see supplemental Methods). Using the latter, multiple SNPs remained significant for EA children for persistent (vs. infrequent), and two SNPs, rs7216389 and rs8076131, remained significant for all three classes (vs. infrequent). Our lead SNP rs2305480 remained significant for the persistent and late-onset classes, whereas AA children were still significant for late-onset.
Wheezing and Asthma
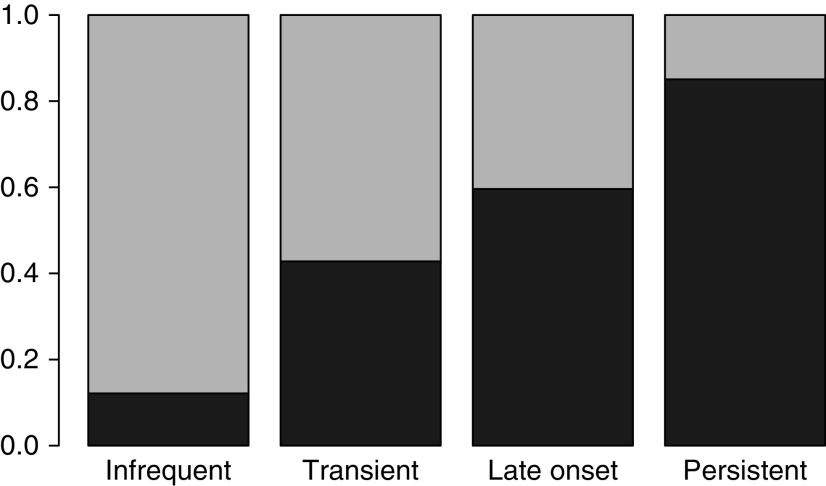
To assess the proportion of subjects with asthma by wheezing group, we classified individuals to their highest probability class. We found substantial variability in the proportion of asthma by class, with 12%, 43%, 60%, and 85% of subjects in the infrequent, transient, late-onset, and persistent wheeze classes, respectively, having a parental report of an asthma diagnosis by a physician by age 11 (Figure 4). Restricting the analysis to individuals with a maximum posterior probability >0.9 (n = 1,914), specifically those individuals with unambiguous wheeze classifications, the proportions with asthma were 15%, 50%, 85%, and 97% for the four classes, respectively. To better understand how so many infrequent and transient wheezers could also have an asthma diagnosis, we looked at the distribution of the age of asthma diagnosis as reported by the parents. This revealed that a large number of subjects acquired their asthma diagnosis at very young ages (58% by age 3).
Figure 4.
The proportion of latent class analysis subjects with doctor-diagnosed asthma by age 11 in each latent class. Subjects were classified to their highest probability latent class and were considered to have asthma if they had a parental report of an asthma diagnosis by a physician. Dark gray indicates positive for asthma by age 11; light gray indicates no asthma by age 11.
Discussion
By applying LCA to wheezing data from children in seven CREW birth cohorts from across the United States, we confirmed heterogeneity in the temporal expression of wheezing during childhood and identified four wheezing phenotypes, which we labeled infrequent, transient, late-onset, and persistent. These phenotypes are qualitatively similar to the temporal patterns of wheezing reported from analyses of wheeze data from other birth cohorts and have been widely replicated in multiple studies in the past two decades since they were originally described (13, 16–21). Some recent latent class analyses using larger birth cohorts from the United Kingdom and the Netherlands (3, 4) have reported five or six wheezing classes, which also included infrequent and persistent classes, together with one or two transient classes (early and late) and one or two late-onset classes (intermediate and late). Given that the number of latent classes chosen using BIC will typically grow with the sample size (13), those studies appear to represent additional refinements of the four main classes rather than major phenotypic differences.
In contrast to our hypothesis that associations with 17q12-21 SNPs would vary by wheezing phenotype, we found associations between multiple SNPs and all three wheezing classes relative to infrequent wheezers. This was most apparent in EA children, for whom the associations exist across the 17q12-21 region, owing to the extended LD. However, there were two strong genetic association signals in both EA and AA subjects: one with rs2517955, in an intron of PGAP and the promoter of ERBB2, and one with rs2305480 in GSDMB, the gene encoding gasdermin-B. This latter SNP is of particular interest, as it was also the lead SNP to emerge in a recent study by Ober and colleagues (8) looking at genetic associations with childhood onset asthma in both EA and AA children. In addition to the strong association signal, they also found rs2305430 to be the most significant expression quantitative trait locus for GSDMB expression in upper airway epithelial cells. Although the exact function of gasdermin-B is still being determined, it plays a key role in pyroptosis in response to intracellular pathogens (22–24). One potential common risk factor for the three non-infrequent wheezing phenotypes is susceptibility to viral respiratory infections, especially those owing to rhinovirus, which is the most frequent cause of lower respiratory illness after age 1 year (25–28). Our results suggest that such susceptibility may be common to all forms of wheezing in early life.
This is also the first study of LCA wheezing phenotypes and genetic associations in AA children. We chose the included SNPs because they were associated with asthma or gene expression in previous studies (26–28). We suggested that they would also show associations with wheezing phenotypes in EA children—a result we confirmed. We then leveraged the different LD structures between EA and AA children to disentangle the effects of the SNPs and better pinpoint those “driving” the associations. As expected, the marked LD observed in children of European ancestry was greatly reduced in AA children (Figure 3), allowing us to rule out several candidate SNPs within the 17q12-21 region, including rs2941504, rs12936231, rs4065275, rs8069202, and rs3859192, that did not show associations with wheezing in the AAs. Although some of the confidence intervals for the lead SNP, rs2305480, included one in AA children, the AA sample size is smaller than the EA sample. These results mirror the findings of Ober and colleagues (8), who used the same transethnic strategy of using the differences in LD to fine-map associations between 17q12-21 SNPs and childhood asthma. Here, we report for the first time that the same SNP, rs2305480, is associated with all early life wheezing phenotypes, adding confidence to our results and providing clues to the mechanistic pathways through which 17q12-21 SNPs lead to asthma in childhood.
Our conclusions differ from those of Granell and colleagues (6), who looked at associations between 17q SNPs and wheezing latent classes in the United Kingdom–based ALSPAC (Avon Longitudinal Study of Parents and Children) cohort, the only other study to look at such genetic associations. After identifying six wheezing classes in over 7,000 European ancestry children followed from birth to 81 months, they concluded that SNPs at this locus were only associated with the intermediate-onset and Persistent classes (i.e., a higher risk ratio for being in those classes vs. the infrequent class). However, a close examination of their results (Table 2 in Granell and colleagues) suggests our findings are largely compatible. In particular, for both their lead SNP, rs8076131, and our lead SNP, rs2305480, they report relative risk ratios significantly greater than one for all five of their wheeze latent classes (vs. infrequent) except the “transient-early” class, whose confidence intervals included one.
Our data suggest that, despite the fact that the wheeze phenotypes have different natural histories and are known to be associated with different risk factors and long-term clinical and spirometric outcomes during childhood (16), they share common genetic predispositions. Thus, this locus may more appropriately be considered a “wheezing locus” than an “asthma locus.” Because previous studies of the relation of variants in 17q12-21 to asthma likely included children as controls who wheezed in early life but lack a physician diagnosis of asthma, they may have underestimated the effect of this locus on asthma risk. Unexpectedly, we found that 50% of transient wheezers had a parental report of an asthma diagnosis by a physician. This result suggests that a substantial proportion of young children who are diagnosed as having asthma may in fact have a transient condition that will not last beyond the preschool years.
Limitations of this study are those that would be expected when multiple cohorts are pooled for analyses. For example, the seven cohorts ascertained wheeze and covariates differently. The data harmonization procedure was an iterative process that accommodated data that used different questions at varying child ages. Furthermore, some data were not collected by some cohorts and thus could not be harmonized. For example, not all cohorts had information on respiratory viruses or used the same panels of allergen-specific IgEs. The asthma variable used in this study was also limited, as more objective measures such as airway hyperresponsiveness were not available for these children.
However, this study has numerous strengths, including being the first to look at genetic associations with wheezing phenotypes in AA children. The population, which consists of cohorts enrolled across the United States over almost three decades, also represents a range of exposures and risk profiles. There was remarkable consistency in the wheeze patterns detected with previously published studies. Sensitivity analyses confirmed that the classes identified were not driven by a single cohort but were largely consistent across cohorts. Furthermore, the inclusion of a large number of AA subjects, who have different LD patterns than EAs, allowed us to exclude previous candidate variants and better determine which variants are truly associated with wheeze.
In summary, we confirmed and expanded previous analyses identifying common patterns of wheeze in childhood. Furthermore, we report associations for one 17q12-21 SNP (rs2305480) that is associated with all three wheezing phenotypes in both AA and EA children. These findings indicate that the complex genotype–phenotype interactions that underlie the strong childhood onset asthma signal at the 17q12-21 locus are functionally related to the pathophysiology of wheezing.
Supplementary Material
Footnotes
Supported by UG3 OD023282. The CREW cohorts are supported as follows: Cincinnati Childhood Allergy and Air Pollution Study: NIH grants R01 ES11170 and R01 ES019890; Columbia Center for Children’s Environmental Health Cohort: NIH grants P01 ES09600, R01 ES008977, P30ES09089, and R01 ES013163 and Environmental Protection Agency grant R827027; Childhood Origins of Asthma: grants P01 HL070831, U10 HL064305, and R01 HL061879; Epidemiology of Home Allergens and Asthma Study: NIH grant R01 AI035786; Infant Immune Study: grant HL56177; Tucson Children’s Respiratory Study: grant R01 HL132523; and Urban Environment and Childhood Asthma: grants NO1AI25496, NO1 AI25482, HHSN272200900052C, HHSN272201000052I, RR00052, M01RR00533, UL1 RR025771, M01 RR00071, UL1 RR024156, UL1 TR001079, UL1RR024992, and UL1TR000040.
Author Contributions: B.H., G.W., D.B., D.O., J.E.G., C.O., A.L.W., and F.D.M. made contributions to the conception or design of the study. G.K.K.H., R.L.M., R.F.L., D.J.J., D.R.G., G.T.O’C., J.E.G., A.L.W., and F.D.M. contributed patient samples and/or data. B.H. and D.B. oversaw or performed all statistical analyses. C.O. performed genotyping. B.H., G.W., and A.L.W. wrote the initial drafts of the manuscript. D.O., J.E.G., C.O., and F.D.M. read and edited early drafts of the manuscript. All authors reviewed and/or critically revised the manuscript for important intellectual content and provided final approval.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.202003-0820OC on February 3, 2021
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: on behalf of ECHO-CREW
References
- 1.Zahran HS, Bailey CM, Damon SA, Garbe PL, Breysse PN. Vital signs: asthma in children - United States, 2001-2016. MMWR Morb Mortal Wkly Rep. 2018;67:149–155. doi: 10.15585/mmwr.mm6705e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caudri D, Savenije OE, Smit HA, Postma DS, Koppelman GH, Wijga AH, et al. Perinatal risk factors for wheezing phenotypes in the first 8 years of life. Clin Exp Allergy. 2013;43:1395–1405. doi: 10.1111/cea.12173. [DOI] [PubMed] [Google Scholar]
- 3.Henderson J, Granell R, Heron J, Sherriff A, Simpson A, Woodcock A, et al. Associations of wheezing phenotypes in the first 6 years of life with atopy, lung function and airway responsiveness in mid-childhood. Thorax. 2008;63:974–980. doi: 10.1136/thx.2007.093187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oksel C, Granell R, Haider S, Fontanella S, Simpson A, Turner S, et al. STELAR investigators, breathing Together investigators. Distinguishing wheezing phenotypes from infancy to adolescence: a pooled analysis of five birth cohorts. Ann Am Thorac Soc. 2019;16:868–876. doi: 10.1513/AnnalsATS.201811-837OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stein MM, Thompson EE, Schoettler N, Helling BA, Magnaye KM, Stanhope C, et al. A decade of research on the 17q12-21 asthma locus: piecing together the puzzle. J Allergy Clin Immunol. 2018;142:749–764.e3. doi: 10.1016/j.jaci.2017.12.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Granell R, Henderson AJ, Timpson N, St Pourcain B, Kemp JP, Ring SM, et al. Examination of the relationship between variation at 17q21 and childhood wheeze phenotypes. J Allergy Clin Immunol. 2013;131:685–694. doi: 10.1016/j.jaci.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmüller J, Ang W, et al. Australian Asthma Genetics Consortium (AAGC) collaborators. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ober C, McKennan CG, Magnaye KM, Altman MC, Washington C, III, Stanhope C, et al. Environmental Influences on Child Health Outcomes-Children’s Respiratory Research Workgroup. Expression quantitative trait locus fine mapping of the 17q12-21 asthma locus in African American children: a genetic association and gene expression study. Lancet Respir Med. 2020;8:482–492. doi: 10.1016/S2213-2600(20)30011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gern JE, Jackson DJ, Lemanske RF, Jr, Seroogy CM, Tachinardi U, Craven M, et al. The Children’s Respiratory and Environmental Workgroup (CREW) birth cohort consortium: design, methods, and study population. Respir Res. 2019;20:115. doi: 10.1186/s12931-019-1088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linzer DA, Lewis JB. poLCA: an R package for polytomous variable latent class analysis. J Stat Softw. 2011;42:1–29. [Google Scholar]
- 11.Greenacre MJ. Compositional data analysis in practice. Boca Raton, FL: CRC Press LLC; 2018. [Google Scholar]
- 12.Aitchison J. The statistical analysis of compositional data. New York, NY: Chapman and Hall; 1986. [Google Scholar]
- 13.Chen Q, Just AC, Miller RL, Perzanowski MS, Goldstein IF, Perera FP, et al. Using latent class growth analysis to identify childhood wheeze phenotypes in an urban birth cohort. Ann Allergy Asthma Immunol. 2012;108:311–315.e1. doi: 10.1016/j.anai.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemanske RF., Jr The childhood origins of asthma (COAST) study. Pediatr Allergy Immunol. 2002;13:38–43. doi: 10.1034/j.1399-3038.13.s.15.8.x. [DOI] [PubMed] [Google Scholar]
- 15.Wright AL, Taussig LM, Ray CG, Harrison HR, Holberg CJ. The Tucson Children’s Respiratory Study: II. Lower respiratory tract illness in the first year of life. Am J Epidemiol. 1989;129:1232–1246. doi: 10.1093/oxfordjournals.aje.a115243. [DOI] [PubMed] [Google Scholar]
- 16.Martinez FD, Wright AL, Taussig LM, Holberg CJ, Halonen M, Morgan WJ The Group Health Medical Associates. Asthma and wheezing in the first six years of life. N Engl J Med. 1995;332:133–138. doi: 10.1056/NEJM199501193320301. [DOI] [PubMed] [Google Scholar]
- 17.Bacharier LB, Beigelman A, Calatroni A, Jackson DJ, Gergen PJ, O’Connor GT, et al. NIAID sponsored Inner-City Asthma Consortium. Longitudinal phenotypes of respiratory health in a high-risk urban birth cohort. Am J Respir Crit Care Med. 2019;199:71–82. doi: 10.1164/rccm.201801-0190OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sordillo JE, Coull BA, Rifas-Shiman SL, Wu AC, Lutz SM, Hivert MF, et al. Characterization of longitudinal wheeze phenotypes from infancy to adolescence in project viva, a prebirth cohort study. J Allergy Clin Immunol. 2020;145:716–719.e8. doi: 10.1016/j.jaci.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tse SM, Coull BA, Sordillo JE, Datta S, Gold DR. Gender- and age-specific risk factors for wheeze from birth through adolescence. Pediatr Pulmonol. 2015;50:955–962. doi: 10.1002/ppul.23113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse SM, Rifas-Shiman SL, Coull BA, Litonjua AA, Oken E, Gold DR. Sex-specific risk factors for childhood wheeze and longitudinal phenotypes of wheeze. J Allergy Clin Immunol. 2016;138:1561–1568.e6. doi: 10.1016/j.jaci.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurukulaaratchy RJ, Fenn MH, Waterhouse LM, Matthews SM, Holgate ST, Arshad SH. Characterization of wheezing phenotypes in the first 10 years of life. Clin Exp Allergy. 2003;33:573–578. doi: 10.1046/j.1365-2222.2003.01657.x. [DOI] [PubMed] [Google Scholar]
- 22.Chao KL, Kulakova L, Herzberg O. Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein. Proc Natl Acad Sci USA. 2017;114:E1128–E1137. doi: 10.1073/pnas.1616783114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu Y, Jin S, Cheng L, Liu G, Jiang Q. Autoimmune disease variants regulate GSDMB gene expression in human immune cells and whole blood. Proc Natl Acad Sci USA. 2017;114:E7860–E7862. doi: 10.1073/pnas.1712127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Miller M, Broide DH. Chromosome 17q21 genes ORMDL3 and GSDMB in asthma and immune diseases. Adv Immunol. 2017;135:1–52. doi: 10.1016/bs.ai.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Jartti T, Gern JE. Role of viral infections in the development and exacerbation of asthma in children. J Allergy Clin Immunol. 2017;140:895–906. doi: 10.1016/j.jaci.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit LA, Bouzigon E, Pin I, Siroux V, Monier F, Aschard H, et al. EGEA Cooperative Group. 17q21 variants modify the association between early respiratory infections and asthma. Eur Respir J. 2010;36:57–64. doi: 10.1183/09031936.00154509. [DOI] [PubMed] [Google Scholar]
- 27.Loss GJ, Depner M, Hose AJ, Genuneit J, Karvonen AM, Hyvärinen A, et al. PASTURE (Protection against Allergy Study in Rural Environments) Study Group. The early development of wheeze: environmental determinants and genetic susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193:889–897. doi: 10.1164/rccm.201507-1493OC. [DOI] [PubMed] [Google Scholar]
- 28.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.