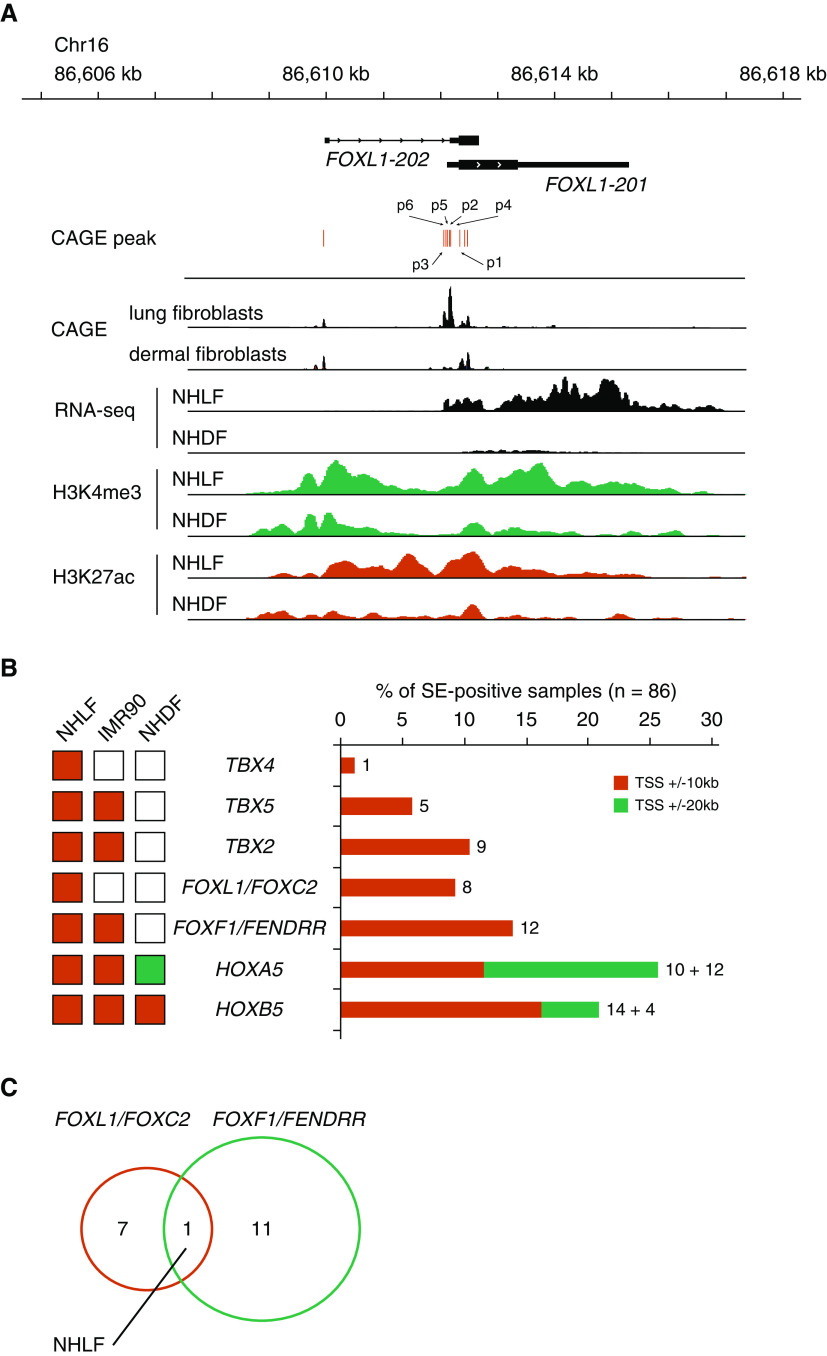
Figure 2.
FOXL1 is associated with super-enhancers in lung fibroblasts. (A) Genomic coordinates, FOXL1 transcripts, and CAGE peaks are indicated. CAGE-sequencing data were merged from three lung fibroblast lines or six dermal fibroblast lines. RNA-sequencing (RNA-seq) signals and ChIP (chromatin immunoprecipitation) peaks of H3K4me3 and H3K27ac in normal human lung fibroblasts (NHLF) or normal human dermal fibroblasts (NHDF) are shown. Note that CAGE peaks, RNA-seq signals, and active histone marks (H3K4me3 and H3K27ac) for FOXL1 are more apparent in lung fibroblasts than in dermal fibroblasts. The same y-axis scales are used for the comparisons between NHLF and NHDF. (B) Left: associations of transcription factors with super-enhancers among NHLF, IMR90, and NHDF. Super-enhancers were identified as reported previously (8). Right: associations of transcription factors with super-enhancers among 86 human cell line and tissue samples. Super-enhancers overlapping with ±10 or 20 kb from the TSSs of the indicated genes were identified. Numbers of samples positive for super-enhancers are indicated. (C) Venn diagram showing that NHLF is the only sample in which both FOXL1/FOXC2 and FOXF1/FENDRR are associated with super-enhancers. Chr = chromosome; FENDRR = FOXF1 adjacent non-coding developmental regulatory RNA; HOXA5 = homeobox A5; SE-positive = super-enhancer–positive; TBX4 = T-box transcription factor 4.