Fig. 4.
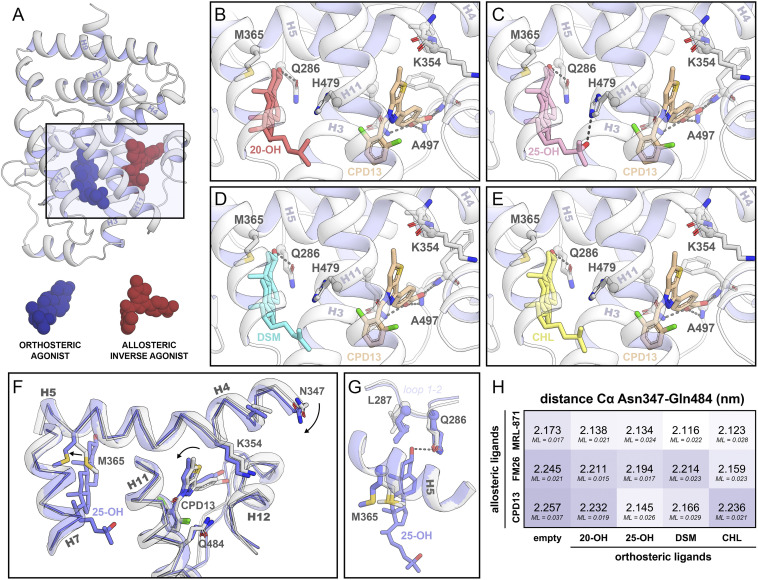
Crystal structures of RORγt in complex with orthosteric and allosteric ligands. (A) Cartoon representation of RORγt in complex with an orthosteric (blue spheres) and an allosteric ligand (red spheres). The rectangle indicates the location of the zoomed-in orthosteric and allosteric LBP. (B–E) The orthosteric and allosteric LBP of RORγt in the presence of various orthosteric ligands (20-OH in red, 25-OH in pink, DSM in blue, and CHL in yellow) and the allosteric ligand compound 13 (brown). (F) Comparison of the crystal structures of RORγt in the presence (blue; PDB entry 6T50) or absence (white; PDB entry 6TLM) of an orthosteric modulator. The presence of the orthosteric modulator shifts helix 4 toward the allosteric pocket, thereby clamping the allosteric ligand. (G) Focused view of the orthosteric LBP. Side chains of Gln286, Leu287, and Met365 are shown for all crystal structures containing ligands in both pockets (12 structures in blue) as well as in absence of an orthosteric modulator (three structures in white; PDB entries 5C4O, 6SAL, and 6TLM). The presence of the orthosteric ligand locks Met365 into a defined state which is conserved for all 12 crystal structures containing orthosteric ligands. (H) Distance (in nm) between the α-carbons of Asn347 (helix 4) and Gln484 (helix 11) in the crystal structures. The maximum-likelihood coordinate error (ML; in nm) is provided for every structure.