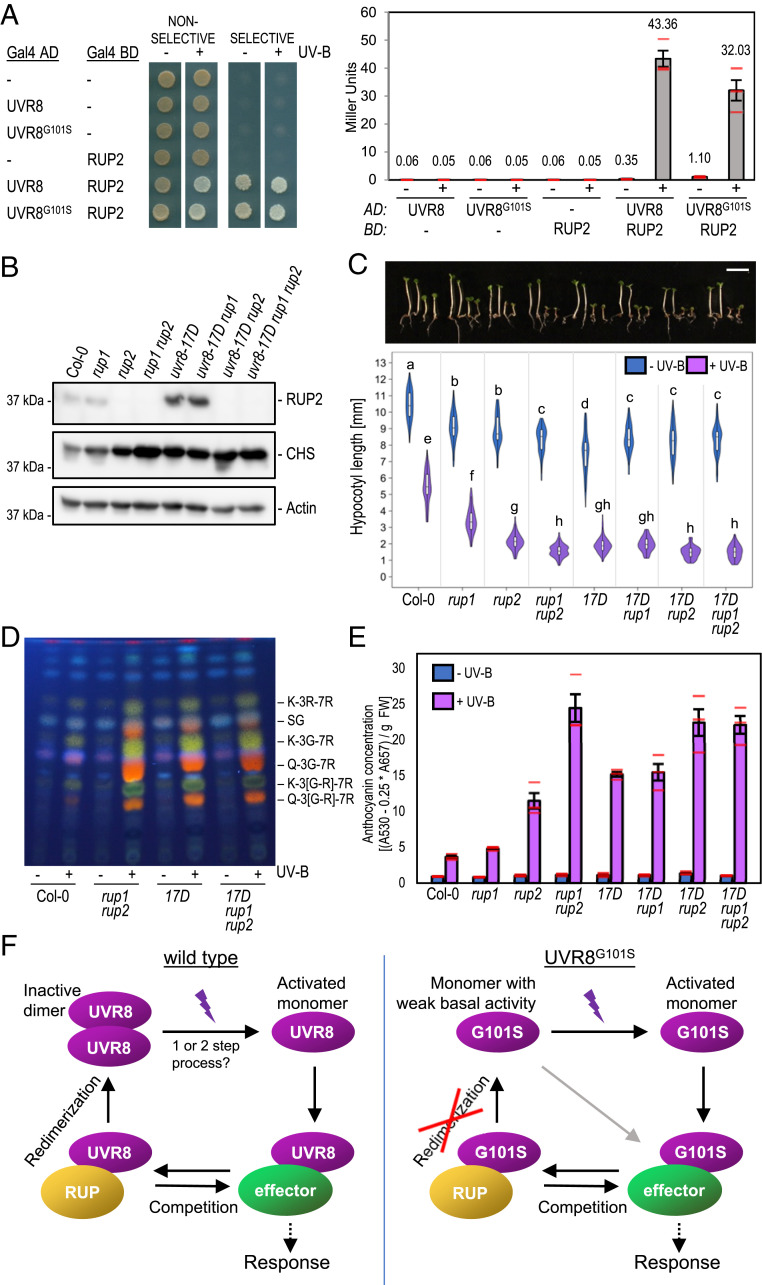
Fig. 5.
The enhanced activity of UVR8G101S depends on the presence of RUP1 and RUP2. (A) Y2H analyses of the interactions of RUP2 with UVR8 and UVR8G101S in the presence or absence of UV-B. Left: growth assay on selective SD/−Trp/−Leu/−His medium. Right: quantitative β-galactosidase assay. AD, activation domain; BD, DNA-binding domain. (B) Immunoblot analysis of RUP2, CHS, and actin (loading control) protein levels in seedlings of wild type (Col-0), rup1, rup2, rup1 rup2, uvr8-17D, uvr8-17D rup1, uvr8-17D rup2, and uvr8-17D rup1 rup2 grown for 4 d in weak white light supplemented with UV-B. (C) Representative images of seedlings described in B (17D, uvr8-17D) and quantification of hypocotyl length (n > 60). Shared letters indicate no statistically significant difference in the means (P > 0.05). (Scale bar, 5 mm.) (D) High-performance thin layer chromatography (HPTLC) analysis of the flavonol glycoside levels in 4-d-old seedlings of wild type (Col-0), rup1 rup2, uvr8-17D, and uvr8-17D rup1 rup2 grown in white light or white light supplemented with UV-B. Identified compounds include: K-3R-7R, kaempferol-3-O-rhamnoside-7-O-rhamnoside; SG, sinapoyl glucose; K-3G-7R, kaempferol-3-O-glucoside-7-O-rhamnoside; Q-3G-7R, quercetin-3-O-glucoside-7-O-rhamnoside; K-3[G-R]-7R, kaempferol 3-O-[rhamnosyl-glucoside]-7-O-rhamnoside; Q-3[G-R]-7R, quercetin 3-O-[rhamnosyl-glucoside]-7-O-rhamnoside. (E) Anthocyanin concentration of seedlings described in B. Values of independent measurements (red bars), means, and SEM are shown (n = 3). (F) Working model of the UVR8 photocycle in the case of wild type (Left) and G101S-mutated UVR8 (Right). In the wild type, UV-B induces monomerization of dimeric UVR8 in a one- or two-step photon absorption process. The activated monomer then interacts with effector proteins such as COP1 and transcription factors to induce a photomorphogenic response. RUP1/RUP2 (RUP) proteins compete with these effectors to abrogate signaling activity. Afterward, RUP-bound UVR8 undergoes redimerization and is brought back to its initial inactive state. UVR8G101S exists as a monomer in vivo and exhibits weak constitutive activity in the absence of UV-B, as seen in overexpression lines. UV-B absorption fully activates the UVR8G101S monomer and RUP proteins still compete with other UVR8 signaling effectors. However, the full cycle cannot be completed because redimerization is intrinsically impossible. This results in sustained UVR8 signaling, leading to enhanced UV-B photomorphogenesis.