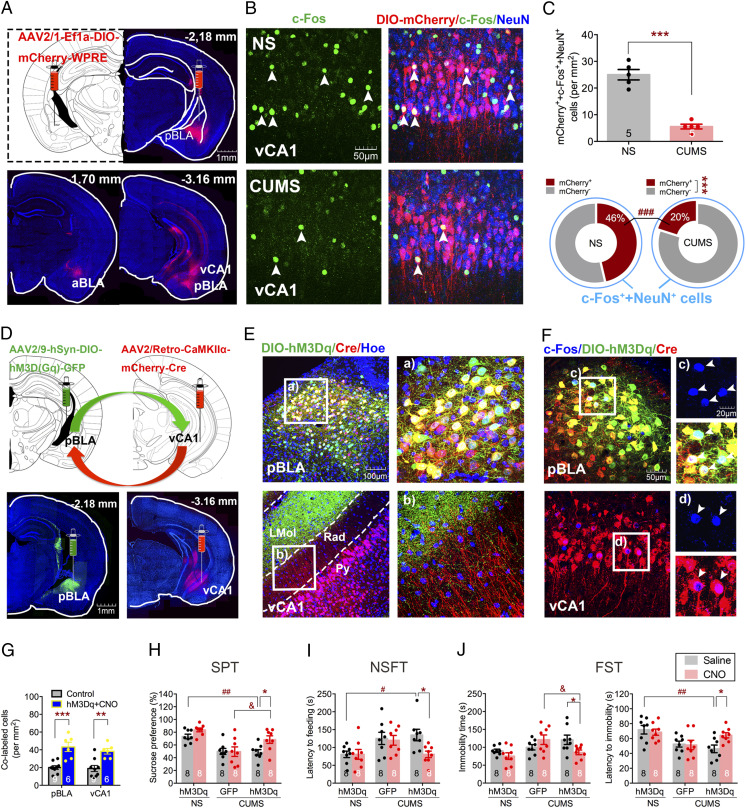
Fig. 2.
pBLA-vCA1 innervation shows a decreased activity in mice subjected to CUMS, and chemogenetic activation of pBLA-vCA1 innervation exerts antidepressant effects. (A) Representative images of Cre-dependent mCherry injection sites in pBLA (red) and mCherry-expressed in aBLA and vCA1 of CaMKIIα-Cre mice. (B) Representative images of DIO-mCherry (red), c-Fos (green), and NeuN (blue) colabeled in vCA1 of NS and CUMS groups, respectively. (C, Top) Quantification of DIO-mCherry, c-Fos, and NeuN triple-positive cells in vCA1 (unpaired Student’s t test, ***P < 0.001). (C, Bottom) Proportion of mCherry-positive or mCherry-negative cells colabeled with c-Fos–positive neurons (two-way ANOVA with Sidak’s post hoc test, ***P < 0.001, mCherry++c-Fos++NeuN+ vs. mCherry−+c-Fos++NeuN+ in CUMS group; ###P < 0.001, mCherry++c-Fos++NeuN+ of CUMS vs. mCherry++c-Fos++NeuN+ of NS group; n = 5 per group). (D) Representative images of DIO-hM3Dq and CaMKIIα-Cre injection sites and expression in pBLA (green; Left) and vCA1 (red; Right), respectively. (E) Representative images of DIO-hM3Dq (green), CaMKIIα-Cre (red) expression, and Hoechst (blue) in pBLA and vCA1, respectively. Rad, stratum radiatum of the hippocampus; Py, pyramidal cell layer of the hippocampus; LMol, lacunosum moleculare layer of the hippocampus. (F, Top) Representative images of c-Fos (blue), DIO-hM3Dq (green), and CaMKIIα-Cre (red) colabeled cells in the pBLA from mice treated with CNO. (F, Bottom) Representative images of c-Fos (blue) and CaMKIIα-Cre (red) colabeled cells in the vCA1 of hM3Dq-expressing mice treated with CNO. White arrowheads indicate c-Fos positive (Top) and colabeled (Bottom) cells. (G) Quantification of colabeled cells in pBLA and vCA1 (unpaired Student’s t test, **P < 0.01, ***P < 0.001). (H) Sucrose preference in SPT. (I) Latency to feeding in NSFT. (J, Left) Immobility time in FST. (J, Right) Latency to immobility in FST (two-way ANOVA with Sidak’s post hoc test, *P < 0.05, #P < 0.05, ##P < 0.01, &P < 0.05). Data are presented as the mean ± SEM.