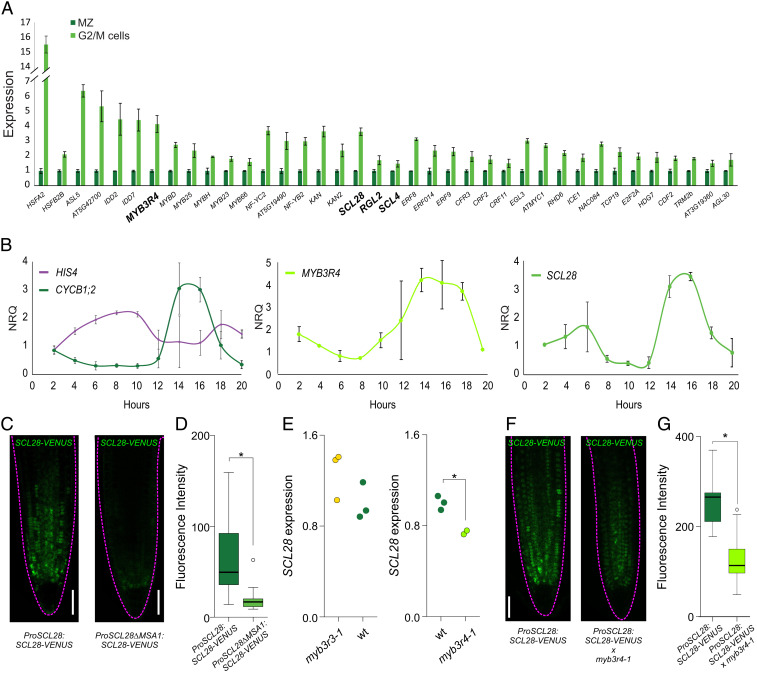
Fig. 2.
SCL28 expression in the MCC. (A) Enrichment of 38 TFs in G2/M cells when compared to complete meristems. Data shown are means ± SE from the three biological replicates obtained in the transcriptome analysis of G2/M cells and complete microdissected root meristems (MZ). All values were normalized to the mean value obtained in complete microdissected root meristems. (B) Time-course expression analysis in cell cycle-synchronized root tips for S and G2/M marker genes (HIS4 and CYCB1;2, respectively), MYB3R4 and SCL28. Expression was estimated by RT-qPCR, and NRQs are reported. Data shown are means ± SE of three biological replicates. (C, Left) Expression pattern of SCL28 determined by LSCM with a ProSCL28:SCL28-VENUS reporter in 6-d-old roots. (C, Right) A reporter with a deletion in one of the MSA elements (ProSCL28ΔMSA1:SCL28-VENUS) was used to analyze the regulation of SCL28 by MYB3R proteins. (Scale bar, 50 μm.) (D) Quantification of fluorescence intensity in the ProSCL28:SCL28-VENUS and ProSCL28ΔMSA1:SCL28-VENUS reporters. Fluorescence intensity in the root meristem was measured in 20 primary transgenics plants transformed with each construct. *P < 0.05 (significant differences; Student’s t test). (E) Expression of SCL28 in wild-type (wt), myb3r3-1, and myb3r4-1 plants. Expression was estimated by RT-qPCR in three biological replicates and normalized to the mean value obtained in wild-type plants. *P < 0.05 (significant differences; Student’s t test). (F) SCL28 expression analyzed by LSCM with a ProSCL28:SCL28-VENUS reporter in 6-d-old wild-type and myb3r4-1 roots. (Scale bar, 50 μm.) (G) Quantification of fluorescence intensity in wild-type and myb3r4-1 roots expressing the ProSCL28:SCL28-VENUS reporter. Fluorescence intensity in the root meristem was measured in 20 roots. *P < 0.05 (significant differences; Student’s t test).