Significance
The transformation of bile acids (BAs) by the gut microbiota is increasingly recognized as an important factor shaping host health. The prerequisite step of BA metabolism is carried out by bile salt hydrolases (BSHs), which are encoded by select gut and probiotic bacteria. Despite their prevalence, the utility of harboring a bsh is unclear. Here, we investigate the role of BSHs encoded by Lactobacillus acidophilus and Lactobacillus gasseri. We show that BA type and BSH substrate preferences affect in vitro and in vivo growth of both species. These findings contribute to a mechanistic understanding of bacterial survival in various BA-rich niches and inform future efforts to leverage BSHs as a therapeutic tool for manipulating the gut microbiota.
Keywords: Lactobacillus, Acidophilus, gasseri, bile salt hydrolase, bile acid
Abstract
Primary bile acids (BAs) are a collection of host-synthesized metabolites that shape physiology and metabolism. BAs transit the gastrointestinal tract and are subjected to a variety of chemical transformations encoded by indigenous bacteria. The resulting microbiota-derived BA pool is a mediator of host–microbiota interactions. Bacterial bile salt hydrolases (BSHs) cleave the conjugated glycine or taurine from BAs, an essential upstream step for the production of deconjugated and secondary BAs. Probiotic lactobacilli harbor a considerable number and diversity of BSHs; however, their contribution to Lactobacillus fitness and colonization remains poorly understood. Here, we define and compare the functions of multiple BSHs encoded by Lactobacillus acidophilus and Lactobacillus gasseri. Our genetic and biochemical characterization of lactobacilli BSHs lend to a model of Lactobacillus adaptation to the gut. These findings deviate from previous notions that BSHs generally promote colonization and detoxify bile. Rather, we show that BSH enzymatic preferences and the intrinsic chemical features of various BAs determine the toxicity of these molecules during Lactobacillus growth. BSHs were able to alter the Lactobacillus transcriptome in a BA-dependent manner. Finally, BSHs were able to dictate differences in bacterial competition in vitro and in vivo, defining their impact on BSH-encoding bacteria within the greater gastrointestinal tract ecosystem. This work emphasizes the importance of considering the enzymatic preferences of BSHs alongside the conjugated/deconjugated BA–bacterial interaction. These results deepen our understanding of the BA–microbiome axis and provide a framework to engineer lactobacilli with improved bile resistance and use probiotics as BA-altering therapeutics.
The human gastrointestinal (GI) tract microbiota plays a critical role in the establishment and maintenance of human health. There is a diversity of microbes that encode millions of genes absent from the human host (1, 2), which are responsible for unique biochemical transformations that occur in the GI tract (3, 4). Perturbations to the gut microbiota and metabolome are associated with the onset and progression of many diseases, such as inflammatory disorders (5–7), metabolic syndromes (8–10), cancers (11–13), and Clostridioides difficile infection (14, 15). Bile acids (BAs), a major component of bile, represent an important class of metabolites that shape host physiology, metabolism, and the gut microbiota (16, 17). BAs are produced by host hepatocytes from cholesterol, stored in the gall bladder, and excreted into the proximal small intestine to aid in the solubilization and absorption of lipophilic nutrients and vitamins after the ingestion of food (18, 19). As they transit through the small intestine, the majority of BAs (>95%) are reabsorbed in the terminal ileum and undergo enterohepatic recirculation, where a portion of BAs escape and reach peripheral organs (19). Remaining BAs that travel to the large intestine are either passively absorbed by the epithelium or excreted in the feces. BA receptors, such as the nuclear receptor farnesoid X receptor or the G protein-coupled bile acid receptor 1 (GPBAR1, also known as TGR5) are expressed throughout the body and are especially abundant in the liver and the GI tract (19–21). BA receptors recognize BAs as agonists or antagonists for receptor signaling and monitor local BA levels to control many aspects of energy metabolism (22), immunity (23, 24), homeostasis (25, 26), and negatively regulate BA synthesis (20, 21).
BAs are weak acids with detergent-like properties that inherently restrict the growth of select bacteria while enriching for others (27–30). Given the dynamic flux of bile through the GI tract, members of the microbiota have evolved strategies to modify BAs, thereby detoxifying BAs and promoting survival (17, 29, 31). Host-derived BAs, referred to as “primary BAs,” are conjugated via an amide bond to either a glycine or taurine (Fig. 1A and SI Appendix, Fig. S1). Conjugated BAs are excreted into the proximal small intestine where they are recognized and cleaved by bacterial bile salt hydrolases (BSHs), which liberate the conjugated amino acid and yield deconjugated BAs (31). This step is considered to be a gatekeeper of downstream BA metabolism because deconjugated BAs serve as precursors for microbial-encoded transformations that generate “secondary BAs” (32, 33), which greatly expand the chemical diversity of the BA pool (34). Secondary BAs that undergo enterohepatic recirculation are recycled and reconjugated in the liver before excretion into the GI tract (19). These conjugated secondary BAs also serve as substrates for BSHs, and once deconjugated can undergo further biotransformation. BSH activity is therefore required for the formation and upkeep of the BA pool, its many functions, and it is likely a critical mechanism by which many intestinal bacteria shape the GI tract niche they inhabit (26, 35).
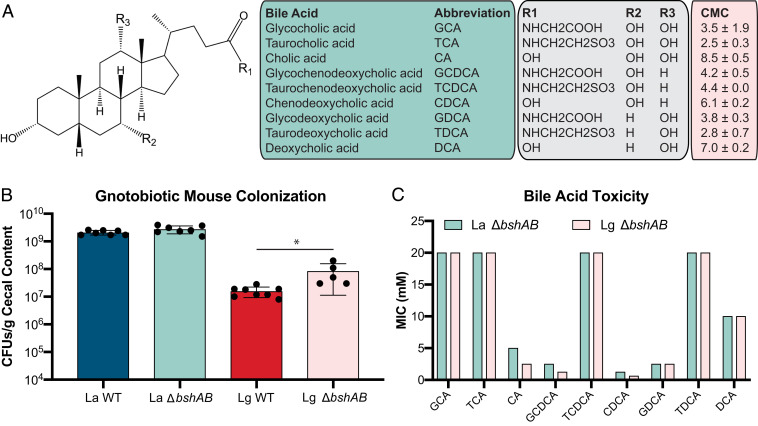
Fig. 1.
BA structures differ in inhibition of Lactobacillus species. (A) BA structures, abbreviations used in this study. All structures shown in SI Appendix, Fig. S1. CMC values calculated in SI Appendix, Fig. S2 and represent the mean CMC from n = 2 experiments ± SEM. (B) WT L. acidophilus (La WT), ΔbshAB L. acidophilus (La ΔbshAB), WT L. gasseri (Lg WT), and ΔbshAB L. gasseri (Lg ΔbshAB) monocolonization of n = 5 to 8 germ-free C57BL/6 mice. CFUs were counted from cecal contents 7 d after colonization. Asterisks represent significant (*P < 0.05) differences from WT by Mann–Whitney U test. (C) BSH-null L. acidophilus and L. gasseri (ΔbshAB) strains were used to determine BA MICs. Bars represent mean MICs from n = 3 independent experiments. MICs did not vary between experiments.
While BSHs are broadly distributed in members of the gut microbiota as well as some pathogens (36–38), many studies have focused on characterizing BSH activity from Lactobacillus species (39–47). Harboring a bsh gene is considered a desirable trait of probiotic lactobacilli due to the dogma that BSH activity contributes to bile resistance and has a hypocholesterolemic effect (39, 48). Recent work has demonstrated that lactobacilli often harbor multiple distinct BSHs as a strategy to adapt to their host niche in the gut, making them a rich source of bsh diversity (44). Additionally, taxonomic profiling of BSHs demonstrated that lactobacilli encode for several prominent and highly active BSH phylotypes (49). Furthermore, lactobacilli are associated with increased BSH function (50). Despite the apparent importance of lactobacilli in the GI tract, the relationship between BSH activity and Lactobacillus host colonization is still not well understood, and the assertion that BSH activity reduces BA toxicity is poorly supported in the literature (38, 51, 52). Additionally, the use of qualitative plate precipitation assays and heterogeneous bile preparations, such as ox-gal, have impeded a mechanistic understanding of Lactobacillus–BSH–BA interactions in vitro and in vivo (53).
Here, we investigate, characterize, and redefine the role of dual BSHs encoded by Lactobacillus acidophilus NCFM and Lactobacillus gasseri ATCC 33323 with respect to bacterial fitness and BA tolerance in vitro and in vivo. Leveraging bacterial genetics and biochemical approaches, we describe how key differences in BSH substrate preferences impact Lactobacillus growth, physiology, and tune the global transcriptional response to BAs. In marked contrast to the previously ascribed functions of BSHs, we report conditions in which BSHs increase the toxicity of BAs. Finally, we show that BSHs can be used to alter microbial dynamics between bsh+ and bsh− lactobacilli. This work reexamines the relationship between BSH-encoding lactobacilli and the BA determinants that mediate colonization of the GI tract and provides a rational basis for the manipulation of BA metabolism that impacts the gut microbiota and host health.
Results
BA Conjugation and Structure Determine Toxicity against Lactobacillus.
L. acidophilus NCK1909 and L. gasseri NCK2253 each encode two BSHs, bshA and bshB. To investigate the contribution of these enzymes to Lactobacillus fitness in vivo, BSH-null strains (ΔbshAB) were constructed by deleting both bsh genes by allelic exchange (54). WT and ΔbshAB strains of L. acidophilus and L. gasseri were monoassociated in germ-free mice for 7 d when ceca were harvested. Given that BSH expression is reported to promote survival in the presence of conjugated BAs (38, 51, 52), we hypothesized that WT strains would better colonize the germ-free GI tract, which lacks microbial BSHs, relative to ΔbshAB strains. Cecal content was enumerated for lactobacilli and the BA pool was quantified using targeted liquid chromatography/mass spectrometry (LC/MS). Evidence of WT bsh expression was exhibited by the increased abundance of deconjugated BAs and the coordinated decrease in conjugated BAs compared to the ΔbshAB mutant (SI Appendix, Fig. S2). While no difference in colonization was detected between L. acidophilus strains, L. gasseri ΔbshAB colonized significantly better than WT (Fig. 1B). This suggests that BSH activity in L. gasseri may be detrimental to the colonization of the GI tract, a finding that contradicts the assumed role of these enzymes (38, 51, 52).
Given this observation, we sought to better define the interaction between Lactobacillus bsh and the glycine or taurine conjugation of three of the most prominent BAs found in the human GI tract: Cholic acid (CA), chenodeoxycholic acid (CDCA), and deoxycholic acid (DCA), illustrated in Fig. 1A. While the BA pool is comprised of many chemically distinct BAs, their structures vary by the mere presence or absence of a hydroxyl or conjugated amino acid. The detergent properties of BAs similarly vary and can induce membrane damage through lipid solubilization, surface protein disruption, or lysis (28). In order to understand the relative detergent properties of BAs, we quantified the critical micelle concentration (CMC) of the BAs used in this study (Fig. 1A and SI Appendix, Fig. S2). In general, conjugation lowered the CMC of a given BA, indicating an increased potential for membrane solubilization and damage (28, 55, 56).
To determine whether BA CMCs correlated with toxicity, the minimum inhibitory concentration (MIC) of each BA was determined for the L. acidophilus ΔbshAB and L. gasseri ΔbshAB mutants to measure the baseline toxicity of individual BAs (Fig. 1C). However, the presence or absence of BSHs did not alter lactobacilli inhibition from deconjugated BAs (SI Appendix, Fig. S4). BSHs are reported to detoxify BAs through deconjugation and our CMC data suggested that the conjugated BAs displayed a greater potential to induce membrane damage, therefore we hypothesized that lactobacilli would display a decreased tolerance for conjugated BAs. Both lactobacilli were resistant to the highest concentrations of glycocholic acid and taurocholic acid (GCA and TCA) tested, yet CA was considerably more toxic relative to its conjugated forms (Fig. 1C). Lactobacillus inhibition from taurochenodeoxycholic acid (TCDCA) and taurodeoxycholic acid (TDCA) was only achieved at high concentrations, whereas glycoochenodeoxycholic acid (GCDCA) and glycodeoxycholic acid (GDCA) were ∼10× more inhibitory. To our knowledge this difference in BA toxicity based on the conjugation and sterol core has never been tested and illustrates that conjugation is an overlooked and potentially important selective pressure exerted by the BA pool on the GI tract microbiota. CDCA was the most inhibitory BA tested, whereas DCA did not potently inhibit either strain. Notably the deconjugation of GDCA to DCA represents the only conversion that outright supports the role of BSHs in detoxifying bile. Additionally, L. acidophilus was twice as resistant to the deconjugated primary BAs CA and CDCA compared to L. gasseri, potentially explaining the lack of a difference between WT and ΔbshAB L. acidophilus in Fig. 1C. Given these MIC results, we concluded that neither the conjugated amino acid nor the CMC of a BA predicted its toxicity, so we hypothesized that BA inhibition of lactobacilli occurs in a manner not limited to the detergent properties of the molecule.
BSH Activity Impacts Lactobacillus Growth in a Substrate-Dependent Manner.
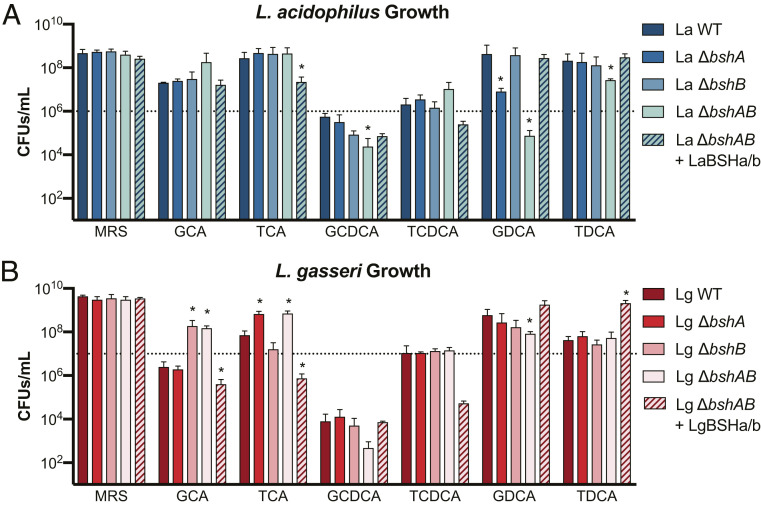
To assess the relative contribution of each bsh to Lactobacillus fitness in vitro, single and double bsh deletion mutants were grown anaerobically in the presence of conjugated BAs and CFUs were enumerated after 24 h (Fig. 2). All conjugated BAs were used within approximate ranges of concentrations found in the GI tract (57). L. acidophilus and L. gasseri growth varied by several orders of magnitude depending on the BA to which they were exposed. WT lactobacilli growth was improved compared to their respective ΔbshAB strains in several conditions tested (L. acidophilus with GDCA and TDCA in Fig. 2A; L. gasseri with GDCA in Fig. 2B). Notably, WT and ΔbshB L. acidophilus growth in the presence of GDCA was robust and comparable to de Man, Rogosa, and Sharpe (MRS) medium alone, while the ΔbshA and ΔbshAB strains were substantially inhibited or killed. Whereas, TDCA elicited a slightly different result by requiring the loss of both L. acidophilus BSHs in order to observe a growth defect. For both L. acidophilus and L. gasseri, WT CFUs were less than the starting CFUs, indicating that there was death over the 24-h experiment. Comparatively, the bsh mutants displayed a larger drop in CFUs demonstrating that BSH activity limited bacterial death when exposed to GCDCA as opposed to limiting growth and highlighting a different strategy by which BSHs can promote lactobacilli survival and presumably host colonization.
Fig. 2.
BSHs impact Lactobacillus fitness in a genotypic and bile acid specific-manner. (A) L. acidophilus and (B) L. gasseri BSH mutants grown for 24 h in MRS, 5 mM GCA, 5 mM TCA, 2.5 mM GCDCA, 2.5 mM TCDCA, 2.5 (A)/1.25 (B) mM GDCA, or 5 mM TDCA. Error bars represent SD from n = 4 independent experiments. Dashed lined denotes the approximate starting CFUs/mL at 0 h. Asterisks represent significant (*P < 0.05) differences from WT by Mann–Whitney U test.
Given that the MICs in Fig. 1C demonstrated that deconjugation could increase BA toxicity, it was plausible that BSH activity impaired lactobacilli growth as well (L. acidophilus with GCA and TCDCA in Fig. 2A; L. gasseri with GCA and TCA in Fig. 2B). Interestingly, the L. gasseri ΔbshA and ΔbshB single mutants exhibited enhanced growth when exposed to TCA and GCA, respectively, hinting that LgBSHa and LgBSHb (bshA and bshB encoded by L. gasseri) have complementary preferences for each type of conjugation. Despite the fact that our findings were limited to a single time point and starting BA per culture, this screen for lactobacilli growth allowed us to broadly examine and identify conditions where BSHs may be beneficial or detrimental. That such a widely penetrant enzyme in the microbiota could be mal-adapting bacteria to GI tract stress is a surprising finding that has yet to be reported. We also investigated whether BSHs could impact Lactobacillus growth in the presence of deconjugated BAs, but no differences were observed (SI Appendix, Fig. S5). Overall, we concluded that the impact of BSH-catalyzed deconjugation on Lactobacillus fitness was context-specific and depended on the conjugated amino acid, the BA core, and the Lactobacillus bsh, which is much more of a dynamic relationship than previously thought.
BSH Activity and Specificity Determines Tolerance to Various BAs.
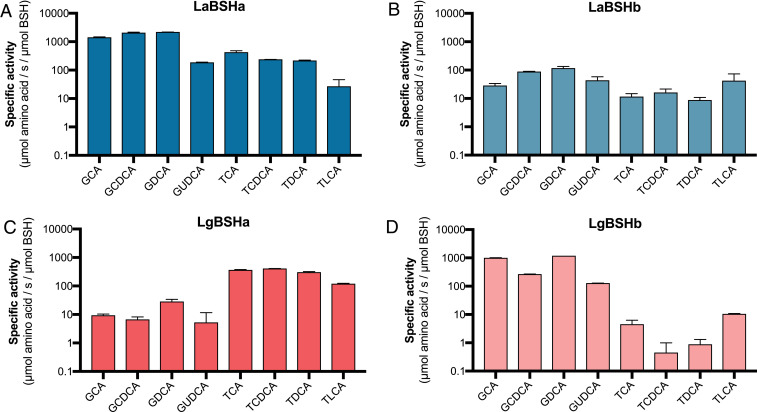
We next hypothesized that differences in substrate specificity could explain why the bshA and bshB of L. acidophilus and L. gasseri varied in their contributions to bacterial fitness observed in Fig. 2. Hereafter, bshA and bshB encoded by L. acidophilus and L. gasseri are referred to as LaBSHa and LaBSHb (57% amino acid identity), and LgBSHa, and LgBSHb (31% amino acid identity), accordingly. LaBSHa, LaBSHb, LgBSHa, and LgBSHb were recombinantly expressed, purified, and assayed for activity on a panel of conjugated BAs using the Ninhydrin reaction to quantify amino acid release (SI Appendix, Fig. S6). All BSHs displayed acidic pH optima, a common feature of these enzymes, and a practical attribute when in acidic environments relevant for lactic acid bacteria (SI Appendix, Fig. S7) (58). Both L. acidophilus BSHs displayed a distinct preference for glycine-conjugated BAs; however, the activity of LaBSHa was ∼10× greater than that of LaBSHb (Fig. 3 A and B), Both BSHs showed preference for GDCA, potentially explaining the large gap in GDCA-supplemented growth between WT L. acidophilus and the ΔbshA or ΔbshAB. The comparatively weaker activity on taurine-conjugated BAs like TDCA supports the need for collective and concerted activity of both LaBSHa and LaBSHb. In contrast, the L. gasseri BSHs LgBSHa and LgBSHb displayed orthogonal functions; LgBSHa preferred taurine-conjugated BAs, whereas LgBSHb was selective for glycine-conjugates (Fig. 3 C and D). These enzymes’ preferences biochemically elucidate the phenotypes of ΔbshA with TCA and ΔbshB with GCA (Fig. 2B), and similarly highlight the benefit of encoding multiple bsh homologs. Despite the patterns between BSH substrate preferences and bsh mutant phenotypes, it is still unknown whether the in vitro BSH activity and substrate preferences are mirrored in bacterial cultures and in vivo.
Fig. 3.
Lactobacillus BSHs display variable preferences for bile acid conjugation. Average specific activities from (A) LaBSHa, (B) LaBSHb, (C) LgBSHa, and (D) LgBSHb were determined by the Ninhydrin assay on the same panel of conjugated BAs used in Fig. 2 supplemented with glycoursodeoxycholic acid (GUDCA) and taurolithocholic acid (TLCA). Error bars represent SD from n = 3 independent experiments.
BSHs are cytoplasmic enzymes that limit the availability of conjugated BAs on which they can act (48). The deconjugated BAs generated inside the cell are either internally sequestered or are disposed of via efflux; however, the localization of their toxic effects has not been defined (48, 59, 60). To investigate whether intracellular BSH compartmentalization is critical for their function or throttles their access to conjugated substrates, equimolar amount of purified BSHs were spiked into the ΔbshAB mutant cultures of L. acidophilus and L. gasseri. Supplementing the lactobacilli ΔbshAB cultures with BSHs functionally complemented or overcomplemented growth of WT in several conditions (see L. acidophilus with GCA, GCDCA, GDCA, TDCA in Fig. 2A; L. gasseri with GCA, TCA, GCDCA, GDCA in Fig. 2B). In other cases, exogenous BSHs distorted ΔbshAB growth or inhibited growth beyond that of WT (TCA in Fig. 2A; TCDCA, TDCA in Fig. 2B). These instances represent conditions in which exogenous BSHs have more access to BAs and are in greater abundance than in WT conditions where BSHs are restricted to the cytoplasm and are produced as a function of Lactobacillus growth. Thus, their activity is artificially inflated. Overall, while we have found that BSHs can display differences in their enzymatic preferences, their cytoplasmic expression can mask the full extent of their selectivity and catalysis.
BSHs Alter Lactobacillus Membrane Physiology.
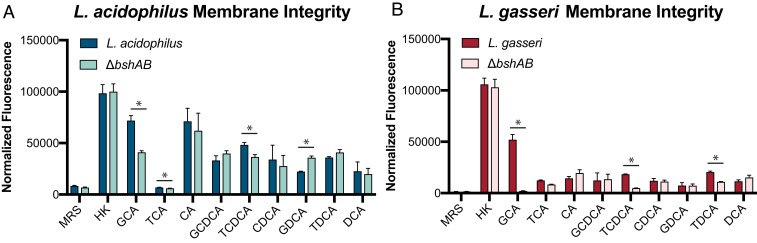
BAs are known to impair bacterial membrane integrity through several means (28). While there was no obvious relationship between the tested BA CMC and MIC for L. acidophilus and L. gasseri (Fig. 1 A and C), this did not rule out that BAs could be damaging membranes and we hypothesized that BSH activity could be curbing or augmenting this damage to the respective benefit or detriment of the bacteria. Using propidium iodide (PI), a membrane impermeant nucleic acid stain commonly used to assess viability, we attempted to describe how BSH activity could impact the integrity and permeabilization of Lactobacillus membranes in the presence of BAs by acutely exposing midlog-grown lactobacilli (61–63).
In several different conditions, L. acidophilus membrane structure was affected by BSH expression (Fig. 4A). GDCA exposure resulted in increased ΔbshAB fluorescence, which is bolstered by the increased killing of the strain compared to WT, but it is unclear how much of the fluorescence is due to permeabilized versus nonviable cells, since the ΔbshAB culture displayed a reduction in CFUs compared to the inoculum after a 24-h culture (Fig. 2A). Exposure to GCA and TCDCA significantly increased the PI fluorescence of WT compared to ΔbshAB. However, this trend opposed the patterns of growth seen in Fig. 2A, revealing that for L. acidophilus, BA-induced membrane damage does not necessarily reflect the molecule’s toxic effects on growth. L. gasseri WT PI fluorescence was greater than ΔbshAB when exposed to GCA (Fig. 4B), which underpins WT inhibition of growth with GCA and possibly death (Fig. 2B). Despite no robust BSH-dependent changes in growth with TCDCA and TDCA, both substrates induced greater membrane damage in WT L. gasseri. Altogether, these results indicate that quantifying membrane damage, rather than growth, may relay more sensitive functional information about BA toxicity in certain cases. These data also demonstrate how BSH expression can regulate lactobacilli membrane integrity and exposure to membrane-damaging BAs, although the exact mechanisms of membrane disruption are unknown and likely vary in a substrate-dependent manner.
Fig. 4.
BSH activity alters membrane integrity and global transcriptome in a BA-specific manner. (A and B) PI staining to assess membrane integrity of midlog (OD 600 nm = 0.7) grown Lactobacillus exposed to various BAs or heat killed (HK). BA concentrations were the same as those used in Fig. 2. Normalized fluorescence was calculated by subtracting background PI fluorescence and normalizing to the starting OD 600 nm at BA exposure. Bars represent average fluorescence from n = 3 independent experiments and error bars represent SD. Asterisks represent significant (*P < 0.05) differences between groups by Welch’s t test.
BSH Expression Alters Lactobacillus Transcriptome during BA Exposure.
Given that both lactobacilli varied in their physiological responses to BA exposure, we wanted to better understand how these bacteria differentially responded to the toxic effects of BAs when equipped with or without a BSH. Previous studies have demonstrated that BA-exposure can lead to transcriptional responses in pathways responsible for membrane organization (64), and we hypothesized that these would be similarly active due to the changes in membrane integrity seen in Fig. 3. We compared whole-transcriptome expression levels for L. acidophilus and L. gasseri during logarithmic growth containing no BAs, CA, GCA, or TCA using RNA sequencing (RNA-seq). While the log2 ratio changes were relatively low, indicating there were not drastic transcriptional changes, there were some significant (P < 0.05) differences depending on the gene or condition. Our data determined that the gene-expression levels for bshA from L. acidophilus was increased similarly (log2 ratio ∼1.10) after growth in all three BAs compared to MRS alone, but there were minimal changes in the expression of bshB (SI Appendix, Fig. S8A). L. gasseri bshA expression was significantly increased in media containing TCA and GCA (log2 ratio 1.11 and 0.80, respectively) and lower expression in CA (log2 ratio –0.66) (SI Appendix, Fig. S8B), indicating positive transcriptional regulation in the presence of conjugated BAs. Interestingly, bshB expression from L. gasseri was significantly decreased after growth in MRS containing CA and TCA (log2 ratio −0.41 and −0.40, respectively) but not GCA, a preferred substrate of this enzyme.
We next determined the differential gene expression for both ΔbshAB mutant and WT after growth with CA, GCA, or TCA compared to MRS alone. Relatively few genes were differentially expressed (SI Appendix, Figs. S8 and S9). L. acidophilus exposure to GCA had the greatest effect on gene expression with 20 genes differentially expressed (SI Appendix, Figs. S8A and S9). Notably, this included genes encoding a transporter, an ABC transporter operon, surface proteins and mnmE and mnmG involved in tRNA regulation, and other cell-surface proteins (SI Appendix, Fig. S8A and Table S1). These cell surface and transporter proteins may contribute to the efflux of the BAs from the cell and are responding to the detergent effect of the BAs.
The exposure of L. gasseri to CA had the largest effect on gene expression, with 8 and 25 genes differentially expressed in the WT and ΔbshAB strains, respectively, indicating a deconjugated BA-specific transcriptomic response (SI Appendix, Figs. S8B and S9 and Table S1). Loss of bshAB resulted in a stronger differential expression for 21 genes after growth in CA (SI Appendix, Figs. S8B and S9), indicating a lack of the bsh genes resulted in an altered transcriptional response to compensate for loss of potential BSH function. Eleven of the 21 genes were up-regulated and 7 of these genes are part of a putative pyrimidine biosynthetic pathway (SI Appendix, Fig. S8B and Table S1). As we have shown CA to be more inhibitory compared to TCA and GCA, it may be expected that the largest stress and transcriptional response was in response to CA. Altogether, our analyses of BA exposure illustrate that the Lactobacillus transcriptional response is weak but pleiotropic, nonetheless, with cell surface proteins being one of the most common to be differentially regulated, although many of their exact functions remain unclear.
BSH-Driven Competition Is Niche- and BA-Specific.
In the GI tract, bacterial competition is an underlying force that shapes the microbiota composition. Competitive strategies for antagonizing or restricting nutrients from competitors have been studied considerably more than those that involve the remediation of environmental stressors, such as BAs. Given that BA biotransformations are a communal activity in the microbiota, we wanted to understand how BSH activity could impact bacterial dynamics and relative fitness between WT and ΔbshAB lactobacilli in coculture. In order to differentiate and enumerate the competing strains, spontaneous rifampicin- and streptomycin-resistant WT and ΔbshAB clones, respectively, were isolated with no detected growth defects on MRS or on select BAs (SI Appendix, Fig. S10). We reasoned that these in vitro competitions could model bacterial interactions in vivo. To further probe these dynamics, exogenous BSHs were also included in cocultures to model the role of extraneous BSHs encoded by other members of the microbiota.
Despite there being favorable conditions for the ΔbshAB mutant over WT L. acidophilus and L. gasseri in monoculture, there was no instance where ΔbshAB displayed greater relative fitness in coculture (Fig. 5 A and B). However, there were several BAs that enriched for WT growth. WT L. acidophilus BSH activity led to ΔbshAB being outcompeted when in the presence of GCDCA and GDCA. These two results mirror the trends from monoculture (Fig. 2A), and show that WT L. acidophilus can use its BSHs to benefit itself, but when toxic deconjugated BAs are generated, competitors are equally damaged. WT L. gasseri dominated cocultures in the presence of GCA and TCA (Fig. 5B), a puzzling result given that in monoculture, WT growth is significantly inhibited by these BAs compared to ΔbshAB (Fig. 2B). Supplementing the competition experiments with exogenous BSHs altered competitive dynamics by neutralizing the fitness advantages of WT lactobacilli, except in the case of WT L. acidophilus with TCA, which unpredictably benefited from the added deconjugation (Fig. 5A). These results indicate that bsh expression can benefit competing lactobacilli, but this advantage is dependent on the type of BA present as well as the existing level of BSH activity.
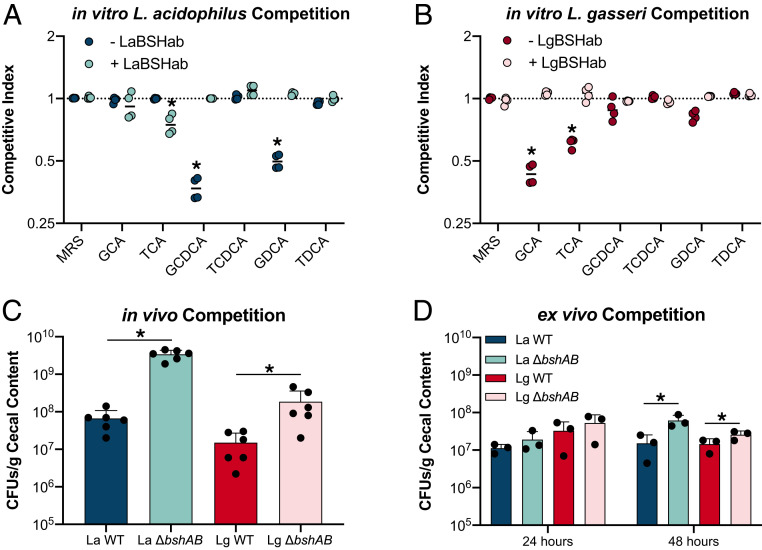
Fig. 5.
BSH activity drives competitive dynamics in vitro and in vivo. (A and B) CIs for Lactobacillus cocultures anaerobically for 24 h in the presence of various BAs. BA concentrations were the same as those used as in Fig. 2. CIs were calculated as follows: CI = Final[Log10(ΔbshAB CFUs)/Log10(WT CFUs)]/Initial[Log10(ΔbshAB CFUs)/Log10(WT CFUs)]. Dashed lines denote a CI = 0. Asterisks represent significant (P < 0.05) differences between groups by Mann–Whitney U test. (C) L. acidophilus and L. gasseri WT/ΔbshAB cocolonization of n = 6 germ-free C57BL/6 mice. CFUs were counted from cecal contents 7 d after colonization. Asterisks represent significant (P < 0.05) differences from WT by Mann–Whitney U test. (D) Cocolonization of n = 3 cecal contents from conventional C57BL/6 mice. Asterisks represent significant (*P < 0.05) differences from WT by unpaired t test.
Our previous observations that BSH activity could be detrimental to lactobacilli fitness did not align with the results of our in vitro competition experiments, so to further understand Lactobacillus BSH-driven competition in a more complex and natural environment, we repeated the germ-free mouse colonization experiment by coassociating mice with WT and ΔbshAB strains of L. acidophilus and L. gasseri (Fig. 5C). We predicted that competing these strains in vivo would more closely approximate bacterial competition in the wild and reveal BSH function in the presence of a dynamic flux of BAs. The bacterial load in the ceca after 7 d of colonization demonstrated that ΔbshAB could compete in the murine GI tract better than WT for both species. While L. gasseri mirrored its trend seen in monoassociated mice, L. acidophilus coassociation better supported the growth of the bsh-null strain (Figs. 1B and 5C). We also tested if the presence of a more complex microbiota and BA pool would change the role of BSH activity during bacterial competition by competing WT and ΔbshAB strains in ex vivo conventional mouse cecal content (Fig. 5D). ΔbshAB strains were more competitive than WT after 48 h of growth. Together, these data demonstrate that BSH activity can drive bacterial competition between strains in an environment-specific fashion, an important consideration for gut colonization and fitness. Additionally, the outcomes from both in vivo gnotobiotic and ex vivo conventional mouse studies support a departure from the dogma dictating that deconjugation is used for BA detoxification.
Discussion
BAs are a diverse group of metabolites that dynamically flux through the GI tract. Their metabolism by resident microbes is a means of cross-talk between the host and its microbiota that has substantial consequences for human health (6, 13, 65–67). Despite the importance of BSHs in initiating the collective metabolism of BAs in the gut, their exact purpose for and impact on the bacteria that encode them is less clear. A large body of work has surveyed the in vitro BSH specificities from many genera of bacteria, especially those encoded by Lactobacillus (58, 68–70), while far fewer studies have genetically assessed the contribution of BSH activity to bacterial fitness (36, 38, 71, 72). Our work has aimed to provide a link between the two by combining genetic and biochemical methods in tandem, whereas early approaches characterizing L. acidophilus BSH activity had lower sensitivity and lacked the same level of resolution (53). Consequently, our findings contradict the purported role of BSHs as enzymes that detoxify BAs and demonstrate that this may be an overgeneralization of their activity in vivo (SI Appendix, Fig. S11A).
Given the complex and pleiotropic effects of BA deconjugation (i.e., shifts in local microbiota composition and function, modulation of intestinal physiology) it is possible that the bile-detoxifying effects of BSHs may be overstated for bsh+ bacteria. Listeria monocytogenes, Listeria innocua, and Brucella abortus all display increased colonization of the murine GI tract and resistance to BA mixtures due to BSH expression (36, 38, 71, 72). It is unclear how or if these bacteria differ from commensal lactobacilli in their response to individual BAs, but perhaps BSH-promoted survival is a trait enriched in pathogenic genera or that works alongside other protective mechanisms to deal with BA stress (60, 64, 73, 74). Nevertheless, our findings show that BSH expression in vivo may come at a fitness cost due to the increase in toxic deconjugated BAs.
While it is known that BAs can restrict the growth of bacteria, we have found that the type of conjugated amino acid dictates much of this toxicity, whereas measuring detergent-like properties (via the CMC) was not predictive of its ability to disrupt bacterial membranes or inhibit Lactobacillus growth (Fig. 1 A and C). These data suggest that the variable levels of glycine:taurine conjugation in bile, for example in different hosts or as a result of diet (75, 76), may play an underappreciated role in shaping the microbiota. Additionally, the glycine/taurine bias that BSHs exhibit may be an evolved trait to alter the levels of glycine/taurine conjugated BA exposure, thereby adjusting the overall toxicity of pooled conjugated and deconjugated BAs. While we chose to focus on prominent BAs found in humans, recent work identifying phenylalanine, tyrosine, and leucine-conjugated BAs in mouse and human GI tracts raise many new questions about the effects of these molecules on members of the microbiota (77).
To our knowledge, our finding that BSH substrate specificity can affect Lactobacillus growth in a conjugation-dependent manner provides evidence explaining why many lactobacilli encode multiple BSHs (Figs. 1C and 2). L. gasseri exemplifies this by correspondingly encoding a BSH specific for glycine and taurine, thereby broadening the substrate range of the strain, whereas the L. acidophilus BSHs seemingly display an additive effect (Fig. 3B). Perhaps an expanded and complementary repertoire of bsh is a competitive feature for intestinal bacteria and a noteworthy signature of gut adaptation that may differ in terms of substrate preferences across hosts with distinct BA pools. This difference in specificity may also have consequences for the host, and previous work has suggested that BSH substrate selectivity can directly modulate host physiology (65). Two recent studies positively or negatively associated specific BSH phylotypes with metabolic and inflammatory diseases that are known to be regulated by BAs (49, 70). Differences in substrate specificity may underlie the protective vs. pathogenic effects of BSHs in the human GI tract or at least serve as a marker for health. Furthermore, whether the bsh-expressing bacteria benefit from establishing these disease niches remains to be seen.
Bacteria have many strategies to cooperate or compete for nutrients and space in the harsh gut environment (78). While BAs are a source of cellular stress that can shape the microbiota, the roles of BA-altering enzymes, such as BSHs, had not been previously tested for their roles in competition. Our in vivo and ex vivo competition studies surprisingly show that both the L. acidophilus and L. gasseri ΔbshAB mutants can outcompete their WT opponents in the gnotobiotic and conventional mouse ceca (Fig. 5 C and D and SI Appendix, Fig. S11B). These findings agree with our observation that WT BSH activity can increase the toxicity of BAs during growth (Fig. 2); however, coculture competitions on individual BAs suggested that BSH could provide a competitive growth advantage under specific conditions (Fig. 5 A and B). We speculate that the murine pool of taurine-conjugated and deconjugated primary BAs may explain the disagreement in these data. Additionally, while we chose not to focus on the murine-exclusive muricholic acids, they may underly some of the unexpected findings in this study and are an important molecule to consider in future studies. That mice somewhat mimic the human BA pool argues for a combined approach using in vivo models as well as in vitro assays supplemented with purified BAs alone or in combination in future studies.
The utility of BSHs as promoters of probiotic colonization and as treatments for disease by reducing serum cholesterol (79), or intervening during obesity (41, 80), is disputed and uncertain. A limited knowledge of the Lactobacillus–BSH–BA relationship, and the enzymatic specificity of a given BSH may explain why the therapeutic success of BSHs and probiotics have had mixed results (81). As next-generation probiotics are engineered using genome-editing tools such as CRISPR-Cas (82), BSHs are promising targets for modification with the potential to improve human health using engineered biotherapeutics. Still, knowledge gaps remain about these enzymes that limit their ability to be used medically. Our findings have provided a foundational understanding of how BSHs equip probiotic lactobacilli to deal with BA stress. Future studies are needed to elucidate the role of BSHs in adapting Lactobacillus and other gut commensals to the competitive environment within the microbiota, and similarly how BSH activity alters the structure and function of that ecosystem.
Materials and Methods
Bacterial Strains and Growth Conditions.
For the construction of bsh mutants, L. acidophilus and L. gasseri strains were grown statically under ambient atmospheric conditions in MRS (Difco Laboratories) broth or glucose semidefined medium (83) at 37 °C or 42 °C. Brain–heart infusion (Difco) broth was used for growth of Escherichia coli strains at 37 °C with aeration. Solid media contained 1.5% (wt/vol) bacteriological agar (Difco) and relevant antibiotics were included in the media where required. Bacterial strain and plasmid details are provided in SI Appendix, Table S2. During experiments, strains were grown statically in a 37 °C Coy anaerobic chamber (5% H2/10% CO2/85% N2). Lactobacillus growth experiments were carried out anaerobically in static MRS media at pH 6.4 for 24 h. Briefly, 2× MRS was diluted with water and aqueous suspensions of BAs (Sigma-Aldrich) to achieve a range of BA concentrations. Endpoint CFUs were determined by diluting cultures in PBS and enumerating on MRS plates. Growth curves were performed in a Tecan plate reader within the anaerobic chamber. Clear flat-bottom plates contained 200 μL of media per well were incubated at 37 °C for 24 h.
DNA Manipulations and Sequence Analysis.
All oligonucleotides used in this study were synthesized by Integrated DNA Technologies (IDT) and are listed in SI Appendix, Table S3. Molecular-grade reagents were purchased from Roche and New England Biolabs. Genomic DNA was isolated using the ZR Fungal/Bacterial DNA miniprep kit (Zymo Research). Plasmid DNA from E. coli was isolated using the QIAprep Spin Miniprep Kit (Qiagen). Chemically competent E. coli EC101 and EC1000 (SI Appendix, Table S2) cells were prepared as described previously (84), and electrocompetent Lactobacillus cells were prepared as described by Walker et al. (85). PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen), QIAquick PCR Purification Kit (Qiagen), Monarch PCR & DNA Cleanup Kit (New England Biolabs), or Monarch DNA Gel Extraction Kit (New England Biolabs). Genewiz performed DNA Sanger sequencing.
Deletion of bsh Genes from L. acidophilus and L. gasseri.
Strains of L. acidophilus and L. gasseri with deletions in a single bsh gene (ΔbshA or ΔbshB) and deletions in both bsh genes (ΔbshAB) were constructed using the upp-based counterselective gene replacement system (54). For each targeted bsh gene the flanking regions (FRs) were amplified by PCR. The FRs for L. acidophilus bshA (LBA0892) were amplified with PCR primer pairs La_BshA_SOEA_F/R (678 bp) and La_BshA_SOEB_F/R (624 bp) (SI Appendix, Table S2). PCR primer pairs La_BshB_SOEA_F/R (665 bp) and La_BshB_SOEB_F/R (743 bp) were used to amplify the FRs for bshB (LBA1078) (SI Appendix, Table S2). For L. gasseri the FRs for bshA (LGAS_00260) were amplified with PCR primer pairs Lg_BshA_SOEA_F/R (749 bp) and Lg_BshA_SOEB_F/R (663 bp) and Lg_BshB_SOEA_F/R (755 bp) and Lg_BshB_SOEB_F/R (669 bp) for bshB (SI Appendix, Table S2). Each FR for each bsh gene was purified and PCR products joined by splicing using overlap extension (SOE) PCR. Each SOE product was digested with SacI and BamHI, purified and ligated into the SacI and BamHI sites of pTRK935. Ligation mixtures were transformed into E. coli EC101 or EC1000. Potential transformants were screened with primer pair upp_ScF and upp_ScR (SI Appendix, Table S3). Sequence integrity of the plasmid insert was confirmed by Sanger sequencing. Plasmids were designated pTRK1123 (FRs of bshA in L. acidophilus), pTRK1124 (FRs of bshB in L. acidophilus), pTRK1206 (FRs of bshA in L. gasseri), and pTRK1207 (FRs of bshB in L. gasseri) (SI Appendix, Table S2). To construct strains with a single bsh deletion in L. acidophilus, plasmids pTRK1123 or pTRK1124 were electroporated into NCK1910, which harbors the helper plasmid pTRK669 that provides repA in trans for the replication of pTRK1123 and pTRK1124. Procedures to isolate and confirm plasmid-free recombinants after targeted plasmid integration into the chromosome and double-cross-over recombination were performed as described previously (54). The confirmed mutant strains with a deletion in bshA or bshB were designated L. acidophilus NCK2521 and NCK2522, respectively. To create a double-deletion mutant in bshA and bshB, the helper plasmid pTRK669 was electroporated into NCK2521. Subsequently, pTRK1124 was electroporated into NCK2521 harboring pTRK669 to facilitate deletion in bshB using the same protocols described in Goh et al. (54), resulting in strain L. acidophilus NCK2523 with a deletion in both bshA and bshB. Single deletions in bshA or bshB and deletions in both bsh genes (ΔbshAB) were similarly made for L. gasseri using NCK2254 as the base strain with plasmids pTRK1206 and pTRK1207 resulting in L. gasseri NCK2678 (ΔbshA), L. gasseri NCK2679 (ΔbshB), and L. gasseri NCK2680 (ΔbshAB).
RNA Isolation.
Total RNA was isolated from L. acidophilus NCK1909 (WT), NCK2523 (ΔbshAB), and L. gasseri NCK2253 (WT) and NCK2680 (ΔbshAB) grown to midlog phase (OD 600 nm ∼0.7) in MRS containing no BA, CA (0.3125 mM), TCA (1.25 mM), or GCA (1.25 mM). Cells were harvested from two biological replicates for each condition by centrifugation (10 mL, 1,717 × g for 10 min at room temperature), pellets flash frozen and stored at −80 °C. Total RNA was extracted using Tri-Reagent (Life Technologies) and purified with the Zymo Direct-Zol RNA Miniprep Kit (Zymo Research). Briefly, the cell pellets were resuspended in 1 mL of Tri-Reagent, added to a screw cap tube containing beads (0.1-mm glass beads, Bio-Spec) and bead-beated for 5 min (five times each for 1-min intervals with 1 min on ice after each interval). Samples were then left at room temperature for 5 min and subsequently centrifuged at 16,873 × g for 20 min at 4 °C. Total RNA was purified using the Zymo Direct-Zol RNA Miniprep Kit (Zymo Research) according to the manufacturer’s instructions. DNA was removed by incubating samples with Turbo DNase as described by the manufacturer (Ambion), purified using the RNA Clean and Concentrator kit (Zymo Research), and checked for integrity by capillary electrophoresis on the Agilent Bioanalyzer (Agilent Technologies). Samples were confirmed to be DNA-free by PCR with L. acidophilus NCFM and L. gasseri ATCC33323 gene-specific primers.
RNA-Seq and Transcriptional Analysis.
Library preparation and sequencing was performed at The High-Throughput Sequencing and Genotyping Unit of the Roy J. Carver Biotechnology Center, University of Illinois at Urbana–Champaign. The Ribozero Bacteria kit (Illumina) was used to remove rRNA followed by library preparation with the TruSeq Stranded RNA Sample Prep kit (Illumina). Libraries were then quantitated via qPCR and sequenced on one lane for 101 cycles from each end of the fragments on a NovaSEq. 6000 using a using a NovaSeq SP reagent kit; reads were 100 nt in length. Fastq files were generated and demultiplexed with the bcl2fastq v2.20 Conversion Software (Illumina). Adapter sequences were removed and raw sequences assessed for quality using Fast QC v0.11.9. Subsequent processes were performed with Geneious Prime 2020.0.5 using default settings. RNA sequence reads for L. acidophilus NCK1909 and NCK2523 were mapped to the L. acidophilus NCFM genome (NC_006814). L. gasseri NCK2253 and NCK2680 sequence reads were mapped to the L. gasseri ATCC 33323 genome (NC_008530). Expression levels were calculated based on the normalized transcripts per million and differential expression was performed with DESeq2 in Geneious. Genes were considered differentially expressed when they had a log2 ratio ≤ −2 or ≥ 2 and a P < 0.05, unless otherwise stated.
BA Critical Micelle Concentrations.
CMCs were determined for the BAs used in this study using Optimizer blueBALLS (G-Biosciences) according to the manufacturer’s instructions and Devlin et al. (63). Eight 1.5-mL Epi tubes containing 250 μL of BA ranging from 1 to 14 mM and two Optimizer blueBALLS were set up in duplicate and were statically incubated for 16 h at room temperature with intermittent vortexing. To read the reactions, tubes were centrifuged for 5 min at 6,000 × g and supernatants were removed and centrifuged at the same speed once more. Of the supernatants, 200 μL were removed, added to clear flat-bottom 96-well plates, and were read at 630 nm in a Tecan Infinite F200 Pro plate reader. The amount of solubilized dye in the supernatant is related to the quantity of BA micelles present. Absorbance values were plotted using GraphPad Prism and were fitted to a five-parameter logistic curve, where the computed inflection point represented by the Log10EC50 corresponds to the CMC (86).
Minimum Inhibitory Concentrations.
BA tolerance measured by MIC was adapted from the bile tolerance assay by Jacobsen et al. (87). Overnight lactobacilli cultures (107 to 108 CFUs/mL) were inoculated 1% into MRS containing a range of BA concentrations. Cultures were anaerobically incubated for 24 h at 37 °C. Following incubation, cultures were serially diluted in PBS and plated on MRS agar to determine if the concentration of BA tested inhibited growth relative to the starting inoculum.
Animals and Housing.
Animal experiments were conducted in the Laboratory Animal Facilities located on the North Carolina State University (NCSU) College of Veterinary Medicine (CVM) campus. The animal facilities are equipped with a full-time animal care staff coordinated by the Laboratory Animal Resources division at NCSU. The NCSU CVM is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. The Institutional Animal Care and Use Committee at the NCSU CVM approved this study. Trained animal handlers in the facility fed and assessed the status of animals several times per day.
Gnotobiotic Mouse Colonization.
Five- to 8-wk-old germ-free C57BL/6J mice (male and female) were challenged once with 109 CFUs of Lactobacillus via oral gavage. Seven days postgavage, animals were humanely killed via CO2 asphyxiation. Cecal contents were collected and a portion was immediately frozen in liquid N2 for downstream metabolomics and in parallel a sample was freshly diluted 1:10 (wt/vol) in sterile PBS. The diluted contents were serially diluted and plated onto MRS to enumerate lactobacilli.
BA Metabolomics.
BA analysis was performed with a UPLC–electrospray ionization (ESI)–MS/MS consisting of a Vanquish UPLC system (Thermo Fisher Scientific) coupled with an Altis TSQ triple quadrupole mass spectrometer (Thermo Fisher Scientific) equipped with a heated ESI source, which enabled the measurement of 20 BAs using the Biocrates BAs kit (Biocrates Life Sciences). To ensure accuracy and precision, the method provides seven calibration standards, a mixture of nine isotope-labeled internal standards, and three quality-control samples. For cecal samples, 0.5-mm ceramic beads (Omni International) were added to 2.0-mL centrifuge tubes containing 50 to 250 mg of sample. Three times the sample volume of extraction buffer containing 20 mM phosphate buffer and ethanol was added to each sample. The samples were homogenized using a genie disrupter (Scientific Industries) for 15 min after vortex mixing for 30 min. Samples were then centrifuged at 15,000 rpm and 4 °C for 10 min and then stored at −80 °C awaiting further processing. On the day of data acquisition, samples were thawed to room temperature and another three times the sample volume of extraction buffer was added. Samples were subjected to vortex mixing for 10 min followed by centrifugation at 15,000 rpm and 4 °C for 15 min. The supernatant was removed and used for analysis. Ten microliters of plasma or extracted fecal, cecal, or ileal matter was added together with 10 μL of the internal standards mixture onto filter spots suspended in the wells of a 96-well filter plate (PALL AcroPrep, PTFE 0.2 μm) fixed on top of a deep-well plate and extracted with 100 μL methanol by shaking at 600 rpm for 20 min. Elution of the methanol extracts was performed using a positive-pressure manifold (Waters) into the lower receiving deep-well plate, which was then detached from the upper filter plate. After adding 50 μL Milli-Q water to the extracts and shaking briefly (600 rpm, 5 min), the sample plate was analyzed by UPLC-MS/MS. All target BAs were baseline-separated using a Restek Raptor C18 column (1.7 mm, 2.1 × 50 mm). Mobile phase A was 5 mM ammonium acetate and mobile phase B was 1:1 methanol/acetonitrile. The gradient program initially started at 0.5 mL/min 35% B with a stepped gradient to 55%B over 4.6 min. The flow rate then increased to 0.8 mL/min at 85%B for an additional 2 min followed by re-equilibration at initial conditions for a total run time of 8.5 min. Samples, standards, blanks and quality controls were analyzed (20-mL injections) in negative ion mode (spray voltage 2.5 kV, ion transfer tube temperature 325 °C, vaporizer temperature 350 °C, sheath gas 50 a.u., aux gas 10 a.u., sweep gas 1 a.u.). MS/MS acquisition was performed in multiple reaction monitoring mode using two specific m/z transitions per analyte. BA concentrations were calculated in QuanBrowser (Thermo Fisher Scientific) and MetIDQ (Biocrates Life Sciences). All calibration curves had R2 values between 0.9911 and 0.9993 and the relative SD (RSD) of internal standards (ITSD) were below 15%.
Recombinant BSH Cloning and Protein Expression.
LaBSHa, LgBSHa, and LgBSHb were PCR-amplified from genomic DNA for cloning into pETite C-His vector (Lucigen) according to the manufacturer’s instructions. Due to poor expression, LaBSHb was codon-optimized and synthesized (IDT). BSH-expressing pETite plasmids were transformed into Rosetta (DE3) pLysS cells (EMD Biosciences) and plated on LB agar containing 30 μg mL−1 kanamycin (Kan) and 20 μg mL−1 chloramphenicol (Cam). After 16 to 20 h of growth at 37 °C, colonies were scraped up and were used to inoculate 1 L of terrific broth plus Kan and Cam for growth at 37 °C. Cultures were grown to an OD 600 nm of ∼0.6 before 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added and cells were grown 30 °C for an additional 16 h. Cells were harvested by centrifugation and cell pellets were flash frozen in liquid N2 until purification.
Recombinant Protein Purification.
Cell pellets were resuspended in 50 mL of lysis buffer (50 mM Na PO4 buffer, 300 mM NaCl, 20 mM imidazole, protease inhibitor tablet [Roche], 10 mM 2-mercapoethanol, pH 8.0) and were lysed by sonication. Lysates were clarified by centrifugation at 25,000 × g for 30 min at 4 °C to remove intact cells. His-tagged BSHs were purified using gravity columns containing 4 mL of fresh HisPur cobalt resin (Thermo Scientific) equilibrated in wash buffer (50 mM Na PO4 buffer, 300 mM NaCl, 20 mM imidazole, pH 8.0). Lysates were allowed to drip through the columns at a rate of ∼1 mL per min. Bound protein was washed using 2 × 20 mL of wash buffer. BSHs were eluted using 10 mL of wash buffer containing 150 mM imidazole and 10 mM DTT. Enzymes were immediately flash frozen in liquid N2 to prevent further oxidation. Enzyme concentrations were quantified using Qubit Protein Assay Kit and protein purity was assessed using 10% or 4 to 20% SDS/PAGE gels (Thermo Scientific).
BSH Activity Assays.
Enzymatic assays were set up according to a previously described two-step method (88). Enzymatic reactions were carried out at 37 °C in 50-μL volumes containing 0.1 M sodium phosphate pH 6.0, 10 mM DTT, 9 mM BA, and 1 to 10 nM BSH. Reactions were quenched using an equal volume of 15% (wt/vol) trichloroacetic acid and were centrifuged at 12,000 × g for 2 min at room temperature to pellet precipitate. To determine the quantity of released amino acid by ninhydrin reaction, 25 μL of the quenched reaction was added to 475 μL of Ninhydrin buffer (0.3 mL glycerol, 0.05 mL of 0.5 M citrate buffer pH 5.5. 0.125 mL of 0.5 M citrate buffer pH 5.5 with 1% [wt/vol] Ninhydrin). The Ninhydrin reaction was developed by boiling for 14 min and cooled on ice for 3 min. A standard curve of glycine or taurine was prepared for each assay. Absorbance at 570 nm was measured in a clear flat-bottom plate in a Tecan Infinite F200 Pro plate reader.
PI Staining.
WT and ΔbshAB strains of L. acidophilus and L. gasseri were captured at midlog (OD 600 nm = 0.8), then back-diluted to a final OD 600 nm = 0.1 into MRS containing BAs for 3 h. Following exposure, bacteria were stained with PI using slight modifications to a previous method (62, 63). Bacteria were heat-killed at 80 °C for 10 min as positive controls for PI staining. Sub-MIC concentrations of deconjugated BAs were included to control for strain-dependent differences in membrane permeabilization. PI fluorescence was measured from flat-bottom clear plates (excitation: 540 nm; emission: 610 nm) and was normalized to OD 600 nm.
Coculture Competition Assays.
In vitro competition experiments were performed identical to monoculture growth assays. MRS agar containing 100 μg/mL of rifampicin and 500 μg/mL of streptomycin were used to select for WT (RifR) and ΔbshAB (StrR) strains, respectively (89, 90). Competing bacteria were inoculated 1:1 and competitive indexes (CI) were used to present relative fitness across conditions. CI was calculated as:
Ex vivo competition experiments were performed in triplicate by inoculating cecal content with 105 CFUs of WT (RifR) and ΔbshAB (StrR) strains/gram of content. Lactobacillus Selection Agar (BD) supplemented with rifampicin or streptomycin was used to select for and enumerate the test strains separately. Cecal contents were harvested from 5 to 8 wk-old WT C57BL/6J mice (Jackson Laboratory) as described above in the gnotobiotic mouse colonization methods.
Supplementary Material
Acknowledgments
We thank DuPont Nutrition and Health for support, and the staff at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana–Champagne. M.H.F. is supported by University of North Carolina Center for Gastrointestinal Biology and Disease postdoctoral fellowship training Grant T32DK07737. This research was supported, in part, by NIH Pilot Grant P30 DK034987 through the University of North Carolina Center for Gastrointestinal Biology and Disease. This work was performed in part by the Molecular Education, Technology, and Research Innovation Center at North Carolina State University, which is supported by the State of North Carolina. The funders had no role in this study design, data collection, and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017709118/-/DCSupplemental.
Data Availability.
The RNA-sequence data has been deposited to the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/bioproject (BioProject ID PRJNA639594 and accession numbers SRR12018454–SRR12018485).
References
- 1.Li J.et al.; MetaHIT Consortium , An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Pasolli E., et al., Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings J. H., Macfarlane G. T., Role of intestinal bacteria in nutrient metabolism. JPEN J. Parenter. Enteral Nutr. 21, 357–365 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Wilson I. D., Nicholson J. K., Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs J. P., et al., A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2, 750–766 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lavelle A., Sokol H., Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Franzosa E. A., et al., Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh P. J., et al., An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Lin C. H., Kohli R., Bile acid metabolism and signaling: Potential therapeutic target for nonalcoholic fatty liver disease. Clin. Transl. Gastroenterol. 9, 164 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng X., et al., Bile acid is a significant host factor shaping the gut microbiome of diet-induced obese mice. BMC Biol. 15, 120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGarr S. E., Ridlon J. M., Hylemon P. B., Diet, anaerobic bacterial metabolism, and colon cancer: A review of the literature. J. Clin. Gastroenterol. 39, 98–109 (2005). [PubMed] [Google Scholar]
- 12.Yoshimoto S., et al., Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499, 97–101 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Ma C., et al., Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science 360, eaan5931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theriot C. M., et al., Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theriot C. M., Bowman A. A., Young V. B., Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. MSphere 1, e00045-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann A. F., Hagey L. R., Bile acids: Chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65, 2461–2483 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foley M. H., O’Flaherty S., Barrangou R., Theriot C. M., Bile salt hydrolases: Gatekeepers of bile acid metabolism and host-microbiome crosstalk in the gastrointestinal tract. PLoS Pathog. 15, e1007581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang J. Y., Bile acids: Regulation of synthesis. J. Lipid Res. 50, 1955–1966 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawson P. A., Karpen S. J., Intestinal transport and metabolism of bile acids. J. Lipid Res. 56, 1085–1099 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., et al., Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2, 721–731 (2002). [DOI] [PubMed] [Google Scholar]
- 21.Goodwin B., et al., A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Li T., Chiang J. Y., Bile acid signaling in metabolic disease and drug therapy. Pharmacol. Rev. 66, 948–983 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia W., Xie G., Jia W., Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiorucci S., Biagioli M., Zampella A., Distrutti E., Bile acids activated receptors regulate innate immunity. Front. Immunol. 9, 1853 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verbeke L., et al., The FXR agonist obeticholic acid prevents gut barrier dysfunction and bacterial translocation in cholestatic rats. Am. J. Pathol. 185, 409–419 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Govindarajan K., et al., Unconjugated bile acids influence expression of circadian genes: A potential mechanism for microbe-host crosstalk. PLoS One 11, e0167319 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hofmann A. F., Roda A., Physicochemical properties of bile acids and their relationship to biological properties: An overview of the problem. J. Lipid Res. 25, 1477–1489 (1984). [PubMed] [Google Scholar]
- 28.Begley M., Gahan C. G., Hill C., The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Islam K. B., et al., Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141, 1773–1781 (2011). [DOI] [PubMed] [Google Scholar]
- 30.van Best N., et al., Bile acids drive the newborn’s gut microbiota maturation. Nat. Commun. 11, 3692 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridlon J. M., Kang D. J., Hylemon P. B., Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Ridlon J. M., Harris S. C., Bhowmik S., Kang D. J., Hylemon P. B., Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7, 22–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ridlon J. M., et al., The ‘in vivo lifestyle’ of bile acid 7α-dehydroxylating bacteria: Comparative genomics, metatranscriptomic, and bile acid metabolomics analysis of a defined microbial community in gnotobiotic mice. Gut Microbes 11, 381–404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton J. P., et al., Human cecal bile acids: Concentration and spectrum. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G256–G263 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Sayin S. I., et al., Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Jones B. V., Begley M., Hill C., Gahan C. G., Marchesi J. R., Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 105, 13580–13585 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marchesini M. I., et al., Brucella abortus choloylglycine hydrolase affects cell envelope composition and host cell internalization. PLoS One 6, e28480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dussurget O.et al.; European Listeria Genome Consortium , Listeria monocytogenes bile salt hydrolase is a PrfA-regulated virulence factor involved in the intestinal and hepatic phases of listeriosis. Mol. Microbiol. 45, 1095–1106 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Begley M., Hill C., Gahan C. G., Bile salt hydrolase activity in probiotics. Appl. Environ. Microbiol. 72, 1729–1738 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar R., Grover S., Batish V. K., Bile salt hydrolase (Bsh) activity screening of lactobacilli: In vitro selection of indigenous Lactobacillus strains with potential bile salt hydrolysing and cholesterol-lowering ability. Probiotics Antimicrob. Proteins 4, 162–172 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Joyce S. A., et al., Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc. Natl. Acad. Sci. U.S.A. 111, 7421–7426 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin J., Negga R., Zeng X., Smith K., Effect of bile salt hydrolase inhibitors on a bile salt hydrolase from Lactobacillus acidophilus. Pathogens 3, 947–956 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DiMarzio M., et al., Identification of a mouse Lactobacillus johnsonii strain with deconjugase activity against the FXR antagonist T-β-MCA. PLoS One 12, e0183564 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Flaherty S., Briner Crawley A., Theriot C. M., Barrangou R., The Lactobacillus bile salt hydrolase repertoire reveals niche-specific adaptation. MSphere 3, e00140-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y., et al., Bile salt hydrolase can improve Lactobacillus plantarum survival in gastrointestinal tract by enhancing their adhesion ability. FEMS Microbiol. Lett. 366, fnz100 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Xu F., et al., The complex structure of bile salt hydrolase from Lactobacillus salivarius reveals the structural basis of substrate specificity. Sci. Rep. 9, 12438 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prete R., Long S. L., Joyce S. A., Corsetti A., Genotypic and phenotypic characterization of food-associated Lactobacillus plantarum isolates for potential probiotic activities. FEMS Microbiol. Lett. 367, fnaa076 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Bustos A. Y., Font de Valdez G., Fadda S., Taranto M. P., New insights into bacterial bile resistance mechanisms: The role of bile salt hydrolase and its impact on human health. Food Res. Int. 112, 250–262 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Song Z., et al., Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome. Microbiome 7, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knarreborg A., Engberg R. M., Jensen S. K., Jensen B. B., Quantitative determination of bile salt hydrolase activity in bacteria isolated from the small intestine of chickens. Appl. Environ. Microbiol. 68, 6425–6428 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Smet I., Van Hoorde L., Vande Woestyne M., Christiaens H., Verstraete W., Significance of bile salt hydrolytic activities of lactobacilli. J. Appl. Bacteriol. 79, 292–301 (1995). [DOI] [PubMed] [Google Scholar]
- 52.Moser S. A., Savage D. C., Bile salt hydrolase activity and resistance to toxicity of conjugated bile salts are unrelated properties in lactobacilli. Appl. Environ. Microbiol. 67, 3476–3480 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McAuliffe O., Cano R. J., Klaenhammer T. R., Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 71, 4925–4929 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goh Y. J., et al., Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 75, 3093–3105 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gómez Zavaglia A., Kociubinski G., Pérez P., Disalvo E., De Antoni G., Effect of bile on the lipid composition and surface properties of bifidobacteria. J. Appl. Microbiol. 93, 794–799 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Matsuoka K., Moroi Y., Micelle formation of sodium deoxycholate and sodium ursodeoxycholate (part 1). Biochim. Biophys. Acta 1580, 189–199 (2002). [DOI] [PubMed] [Google Scholar]
- 57.Gilliland S. E., Staley T. E., Bush L. J., Importance of bile tolerance of Lactobacillus acidophilus used as a dietary adjunct. J. Dairy Sci. 67, 3045–3051 (1984). [DOI] [PubMed] [Google Scholar]
- 58.Dong Z., Lee B. H., Bile salt hydrolases: Structure and function, substrate preference, and inhibitor development. Protein Sci. 27, 1742–1754 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kurdi P., et al., Cholic acid is accumulated spontaneously, driven by membrane deltapH, in many lactobacilli. J. Bacteriol. 182, 6525–6528 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Piddock L. J., Multidrug-resistance efflux pumps—Not just for resistance. Nat. Rev. Microbiol. 4, 629–636 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Maurice C. F., Haiser H. J., Turnbaugh P. J., Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152, 39–50 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stiefel P., Schmidt-Emrich S., Maniura-Weber K., Ren Q., Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 15, 36 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Devlin A. S., Fischbach M. A., A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sánchez B., et al., Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187, 5799–5808 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao L., et al., A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7, 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hang S., et al., Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Theriot C. M., Petri W. A. Jr, Role of microbiota-derived bile acids in enteric infections. Cell 181, 1452–1454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chand D., et al., Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta, Gen. Subj. 1861, 2981–2991 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Jarocki P., Targoński Z., Genetic diversity of bile salt hydrolases among human intestinal bifidobacteria. Curr. Microbiol. 67, 286–292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jia B., Park D., Hahn Y., Jeon C. O., Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 11, 1300–1313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Begley M., Sleator R. D., Gahan C. G., Hill C., Contribution of three bile-associated loci, bsh, pva, and btlB, to gastrointestinal persistence and bile tolerance of Listeria monocytogenes. Infect. Immun. 73, 894–904 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Delpino M. V., et al., A bile salt hydrolase of Brucella abortus contributes to the establishment of a successful infection through the oral route in mice. Infect. Immun. 75, 299–305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horáčková Š., Plocková M., Demnerová K., Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol. Adv. 36, 682–690 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Lee K., Lee H. G., Choi Y. J., Proteomic analysis of the effect of bile salts on the intestinal and probiotic bacterium Lactobacillus reuteri. J. Biotechnol. 137, 14–19 (2008). [DOI] [PubMed] [Google Scholar]
- 75.Vessey D. A., The biochemical basis for the conjugation of bile acids with either glycine or taurine. Biochem. J. 174, 621–626 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hardison W. G., Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology 75, 71–75 (1978). [PubMed] [Google Scholar]
- 77.Quinn R. A., et al., Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bauer M. A., Kainz K., Carmona-Gutierrez D., Madeo F., Microbial wars: Competition in ecological niches and within the microbiome. Microb. Cell 5, 215–219 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choi S. B., Lew L. C., Yeo S. K., Nair Parvathy S., Liong M. T., Probiotics and the BSH-related cholesterol lowering mechanism: A Jekyll and Hyde scenario. Crit. Rev. Biotechnol. 35, 392–401 (2015). [DOI] [PubMed] [Google Scholar]
- 80.Li F., et al., Microbiome remodelling leads to inhibition of intestinal farnesoid X receptor signalling and decreased obesity. Nat. Commun. 4, 2384 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suez J., Zmora N., Segal E., Elinav E., The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729 (2019). [DOI] [PubMed] [Google Scholar]
- 82.Hidalgo-Cantabrana C., Goh Y. J., Pan M., Sanozky-Dawes R., Barrangou R., Genome editing using the endogenous type I CRISPR-Cas system in Lactobacillus crispatus. Proc. Natl. Acad. Sci. U.S.A. 116, 15774–15783 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimmel S. A., Roberts R. F., Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp. bulgaricus RR. Int. J. Food Microbiol. 40, 87–92 (1998). [DOI] [PubMed] [Google Scholar]
- 84.Hanahan D., “Techniques for transformation of E. coli” in DNA Cloning: A Practical Approach, Glover D. M., Ed., (IRL Press Ltd., Oxford, UK, 1985), Vol. 1, pp. 109–135. [Google Scholar]
- 85.Walker D. C., Aoyama K., Klaenhammer T. R., Electrotransformation of lactobacillus acidophilus group A1. FEMS Microbiol. Lett. 138, 233–237 (1996). [DOI] [PubMed] [Google Scholar]
- 86.Gottschalk P. G., Dunn J. R., The five-parameter logistic: A characterization and comparison with the four-parameter logistic. Anal. Biochem. 343, 54–65 (2005). [DOI] [PubMed] [Google Scholar]
- 87.Jacobsen C. N., et al., Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65, 4949–4956 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanaka H., Hashiba H., Kok J., Mierau I., Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 2502–2512 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goh Y. J., Klaenhammer T. R., Insights into glycogen metabolism in lactobacillus acidophilus: Impact on carbohydrate metabolism, stress tolerance and gut retention. Microb. Cell Fact. 13, 94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Call E. K., Goh Y. J., Selle K., Klaenhammer T. R., O’Flaherty S., Sortase-deficient lactobacilli: Effect on immunomodulation and gut retention. Microbiology (Reading) 161, 311–321 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-sequence data has been deposited to the Sequence Read Archive database, https://www.ncbi.nlm.nih.gov/bioproject (BioProject ID PRJNA639594 and accession numbers SRR12018454–SRR12018485).