Figure 1.
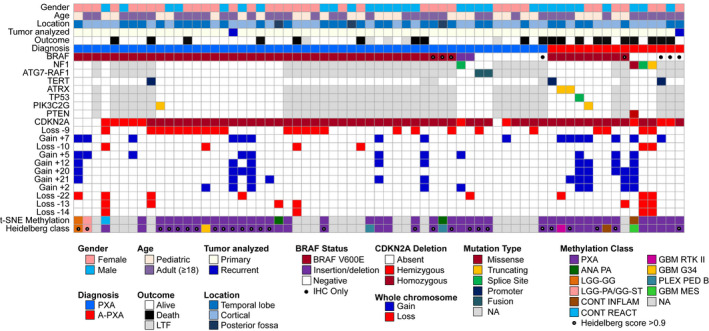
Clinical and molecular characteristics of the patient cohort. BRAF p.V600E mutations were identified by targeted sequencing or (●) immunohistochemistry (IHC) only. TERT promoter mutations were identified by targeted sequencing or ddPCR. CDKN2A/B deletion and whole chromosome gains and losses were derived from MIP data. Genome‐wide methylation data were analyzed against the Heidelberg reference brain tumor cohort (6). Methylation class was defined by unsupervised analysis using t‐SNE dimensionality reduction and utilizing the Heidelberg online classification tool (classifier version v11b4). A calibrated Heidelberg score of >0.9 (●) was considered a definitive match to the reference methylation class. ANA PA, anaplastic pilocytic astrocytoma; LGG‐GG, low‐grade glioma, ganglioglioma; LGG‐PA/GG‐ST, low‐grade glioma, subclass hemispheric pilocytic astrocytoma and ganglioglioma; CONT INFLAM, control tissue, inflammatory tumor microenvironment; CONT REACT, control tissue, reactive tumor microenvironment; GBM RTK II, glioblastoma, IDH wild‐type, subclass RTK II; GBM G34, glioblastoma, IDH wild‐type, H3.3 G34 mutant; PLEX PED B, plexus tumor, subclass pediatric B; GBM MES, glioblastoma, IDH wild‐type, subclass mesenchymal; NA, not available; LTF, lost to follow‐up.
