Figure 2.
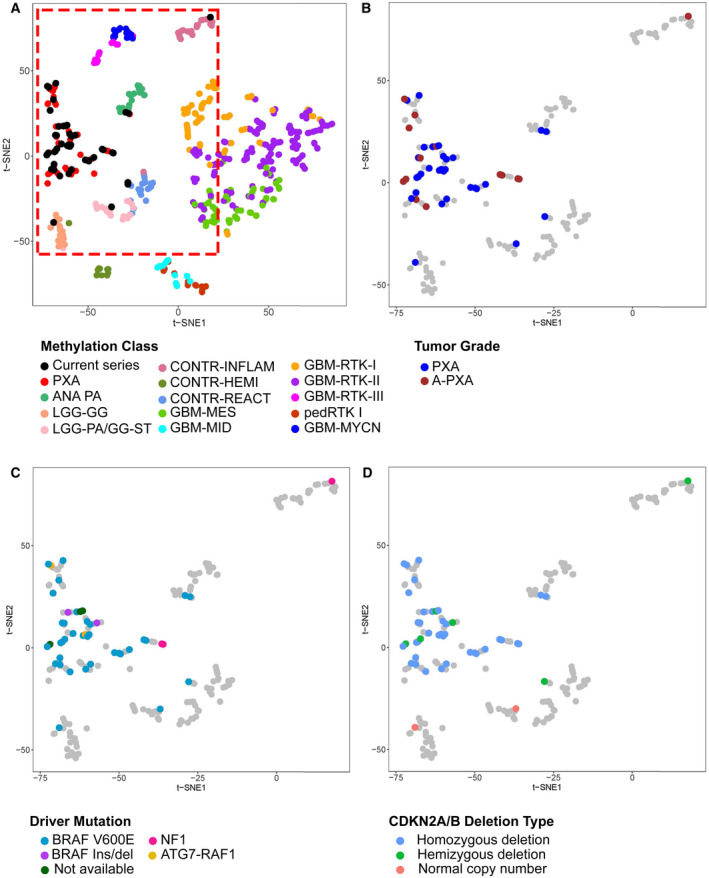
Genome‐wide methylation profiling and methylation‐based classification of PXA. (A) Clustering of PXA relative to published reference methylation classes. Analysis was performed using the top 5000 most differentially methylated probes with t‐SNE visualization. Current series denotes cases from our cohort. (B‐D) Grouping of current cases relative to (B) tumor grade, (C) genetic driver alteration and (D) CDKN2A/B deletion type. Reference cases are shown in grey within the area denoted in red in panel A. Reference methylation classes included: PXA, (anaplastic) pleomorphic xanthoastrocytoma; ANA PA, anaplastic pilocytic astrocytoma; LGG‐GG, low‐grade glioma, ganglioglioma; LGG‐PA/GG‐ST, low‐grade glioma, subclass hemispheric pilocytic astrocytoma and ganglioglioma; CONT‐INFLAM, control tissue, inflammatory tumor microenvironment; CONT‐HEMI, control tissue, hemispheric cortex; CONT‐REACT, control tissue, reactive tumor microenvironment; glioblastoma IDH wild‐type subgroups: GBM‐MES, mesenchymal subclass; GBM‐MID, midline subclass; GBM‐RTK‐I subclass; GBM RTK‐II subclass; GBM‐RTK‐III subclass; pedRTK I subclass; GBM‐MYCN subclass. Two cases analyzed at only recurrence/anaplastic transformation are included with A‐PXA.
