Figure 1.
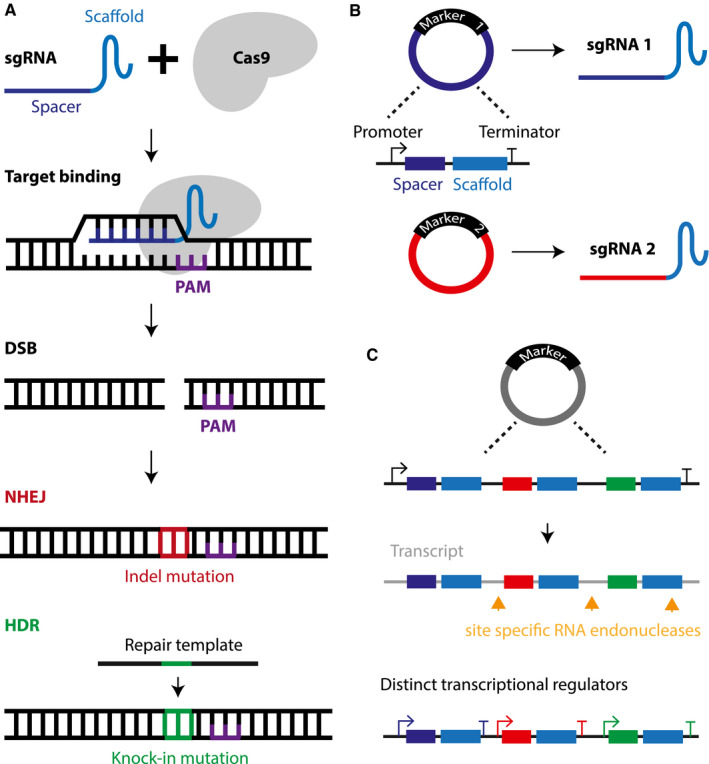
A. The Cas9 nuclease of the clustered regularly interspaced short palindromic repeat (CRISPR) gene editing platform recognizes a DNA sequence of interest via the single guide (sg) RNA that forms a complex with Cas9. The target DNA must contain the protospacer adjacent motif (PAM) sequence for the Cas9‐sgRNA complex to bind and induce a double strand break (DSB). The damaged DNA is then repaired by the non‐homologous end joining (NHEJ) pathway, which is error‐prone and may produce insertion or deletion (Indel) mutations that in turn can cause a gene knockout by the formation of premature stop codons. Alternatively, a precise gene knock‐in can be generated by harnessing the homology directed repair pathway (HDR), which requires a homologous DNA template. B. Multiplexing is facilitated by the short construct size of the plasmid carrying the sgRNA sequence and the various selection markers than can be applied to identify successfully transfected cell populations. C. Multiple sgRNA sequences can be integrated into a single plasmid and the different sgRNA are then delivered via post‐translational processing of the mRNA transcript by site specific RNA endonucleases or distinct promoter sequences for each sgRNA sequence.
