Significance
In this work, we identified the regulatory mechanism of the key control point of cyanobacterial carbon metabolism, the glycolytic phosphoglycerate mutase (PGAM) reaction, converting 3-PGA into 2-PGA and thereby exporting organic carbon from the photosynthetic Calvin cycle. We show that PGAM activity is controlled by a small modulator protein PirC (product of sll0944), which inhibits the enzyme through protein–protein interaction. The availability of PirC for PGAM inhibition is controlled by the pervasive carbon/nitrogen balance regulator PII, which sequesters PirC at low 2-oxoglutarate levels and releases it at high 2-oxoglutarate levels. PirC-mediated inhibition of PGAM triggers glycogen accumulation, and disrupting this regulation allows the redirection of carbon flux, a decisive requirement for transforming cyanobacteria into green factories.
Keywords: glycogen metabolism, polyhydroxybutyrate, cyanobacteria, nitrogen starvation, carbon flow
Abstract
Nitrogen limitation imposes a major transition in the lifestyle of nondiazotrophic cyanobacteria that is controlled by a complex interplay of regulatory factors involving the pervasive signal processor PII. Immediately upon nitrogen limitation, newly fixed carbon is redirected toward glycogen synthesis. How the metabolic switch for diverting fixed carbon toward the synthesis of glycogen or of cellular building blocks is operated was so far poorly understood. Here, using the nondiazotrophic cyanobacterium Synechocystis sp. PCC 6803 as model system, we identified a novel PII interactor, the product of the sll0944 gene, which we named PirC. We show that PirC binds to and inhibits the activity of 2,3-phosphoglycerate–independent phosphoglycerate mutase (PGAM), the enzyme that deviates newly fixed CO2 toward lower glycolysis. The binding of PirC to either PII or PGAM is tuned by the metabolite 2-oxoglutarate (2-OG), which accumulates upon nitrogen starvation. In these conditions, the high levels of 2-OG dissociate the PirC–PII complex to promote PirC binding to and inhibition of PGAM. Accordingly, a PirC-deficient mutant showed strongly reduced glycogen levels upon nitrogen deprivation, whereas polyhydroxybutyrate granules were overaccumulated compared to wild-type. Metabolome analysis revealed an imbalance in 3-phosphoglycerate to pyruvate levels in the pirC mutant, confirming that PirC controls the carbon flux in cyanobacteria via mutually exclusive interaction with either PII or PGAM.
Cellular homeostasis relies on the capacity of living systems to adjust their metabolism in response to changes in the environment. Therefore, organisms must be able to sense the metabolic state and tune it in response to environmental fluctuations. It has been proposed that cyanobacteria do not extensively rely on direct environmental sensing but rather are primarily concerned about their internal metabolic state (1). This “introvert” lifestyle requires that they constantly and precisely monitor their intracellular milieu in order to detect imbalances caused by external perturbations. The maintenance of carbon/nitrogen (C/N) homeostasis is one of the most fundamental aspects of cellular physiology. For photoautotrophic organisms like cyanobacteria, it is essential to tightly interconnect CO2 fixation and nitrogen assimilation. To fulfill this task, cyanobacteria use a sophisticated signaling network organized by the pervasive PII-signaling protein. PII proteins are fundamental for this task in most free-living prokaryotes and chloroplasts of green plants (2). They act as multitasking signal integrators, combining information on the metabolic C/N balance through interaction with the metabolite 2-oxoglutarate (2-OG) and on the cellular energy state by competitive adenosine triphosphate (ATP) or adenosine diphosphate (ADP) binding. 2-OG is ideally suited as a status reporter metabolite for C/N balance, as this tricarboxylic acid (TCA) cycle intermediate represents the precursor metabolite into which ammonia is incorporated through the nitrogen assimilatory reactions catalyzed by the glutamine-synthetase–glutamate-synthase (GS/GOGAT) cycle (3).
The interaction of PII proteins with various effector molecules, the conformational changes that ensue from these interactions, and their perception by the targets have been elaborated in great detail [recently reviewed (3–6)]. The three intersubunit clefts of the trimeric PII proteins contain intercommunicating effector-molecule–binding sites; ADP and ATP compete for occupying these sites, and binding of ATP, but not ADP, creates a coordination sphere for the effector 2-OG through a bridging Mg2+ ion. Depending on the effector molecules bound, the large and flexible target-binding loops (termed T-loops), protruding from the effector binding sites, can adopt specific conformations, allowing signal receptor proteins to read out the metabolic information through protein–protein interactions (5). A variety of key metabolic enzymes, transcription factors, and transport proteins use this signaling path to tune their activity in response to the metabolic state. PII in its different conformations can directly interact with various target proteins such as the N-acetyl-l-glutamate kinase, catalyzing the committed step in arginine biosynthesis (7, 8); the acetyl-CoA carboxylase, catalyzing the rate-limiting step in fatty acid biosynthesis (9); the phosphoenolpyruvate carboxylase, which catalyzes an anaplerotic carbon fixation (10); or the glutamine-dependent nicotinamide adenine dinucleotide (NAD+) synthetase (11). Besides tuning the activity of enzymes, recent analyses revealed that, through direct protein–protein interaction, the abundant PII proteins can also regulate transport activities, including an ensemble of nitrogen transporters such as the nitrate/nitrite transport system, the urea transport system, and the ammonium transporter (12).
A different mechanism underlies the ability of PII proteins to modulate gene expression in response to different C/N ratios. In this case, the effect of PII proteins is mediated through binding to a small signaling mediator protein called PipX (PII-interacting protein X), which acts as a transcriptional coactivator of the global nitrogen control transcription factor NtcA. The latter controls a large regulon of about 80 genes (13). The mediator PipX swaps between PII- and NtcA-bound states, thereby either tuning down or stimulating the activity of NtcA, respectively (6, 14). Partner swapping of PipX occurs in response to the effector molecule 2-OG and the ATP/ADP balance (14).
In a previous PII protein interaction study, several putative PII interactors of unknown function were identified (12). The most prominent hit was the product of the sll0944 gene, a member of the NtcA regulon (11, 13). The sll0944 gene product is annotated in Uniprot (https://www.uniprot.org/uniprot/P77971) as a protein of unknown function. Close homologs are widespread in the cyanobacterial phylum, pointing to an important function of this protein in the cyanobacterial metabolism. We previously observed that sll0944 is up-regulated both at the transcriptional (15) and posttranscriptional (16) level during the response of the model cyanobacterium Synechocystis PCC 6803 (from now on termed Synechocystis) to nitrogen starvation. Following nitrogen depletion, the CO2-fixation products are redirected toward glycogen synthesis, and, concomitantly, the phycobiliproteins and the entire photosynthetic machinery are proteolytically degraded, causing loss of pigments of the cells (referred as chlorosis) (15). Moreover, the carbon polymer polyhydroxybutyrate (PHB) slowly accumulates in granular structures, which are derived from glycogen turnover (17, 18). The metabolic activities decrease as the cells enter into a dormant-like state, in which they can survive for months. As soon as a combined nitrogen source becomes available again, chlorotic cells rapidly awake and resume metabolism (15, 18, 19). This is accompanied by a gradual decrease in the levels of Sll0944 (19).
This study aimed to clarify the role of the Sll0944 protein in Synechocystis and its involvement in PII signaling. Our results indicate that Sll0944 regulates the glycolytic carbon flux in a PII-dependent manner through interaction with the 2,3-phosphoglycerate–independent phosphoglycerate mutase (PGAM) in response to the nitrogen status sensed via 2-OG. Specifically, we found that Sll0944 swaps between PII and PGAM in a 2-OG–dependent manner. This establishes PGAM as a key control point of cyanobacterial carbon flow, as predicted previously by kinetic modeling of the cyanobacterial low-carbon response (20, 21), and Sll0944 as the key regulator of cyanobacterial carbon metabolism. We therefore named the sll0944 product PirC (PII-interacting regulator of carbon metabolism).
Results
In Silico Analysis Reveals High Conservation of Sll0944 (PirC) among Cyanobacteria.
According to the Uniprot database, the gene sll0944 (from now on named PirC) of Synechocystis codes for a 164-amino-acid–long “uncharacterized protein” (https://www.uniprot.org/uniprot/P77971). A databank search for orthologues using the Basic Local Alignment Search Tool (BLAST) revealed that PirC is highly conserved among cyanobacteria. All the homologous proteins contain the Domain of Unknown Function 1830 (http://pfam.xfam.org/family/PF08865). A conserved NtcA-binding site 5′-GTN10AC-3ʹ, which is responsible for nitrogen-starvation–induced expression (22), is situated in front of the respective genes. These findings suggest that pirC and its orthologs might all be responsive to nitrogen starvation. Gene neighboring analysis in 53 cyanobacterial genomes revealed that in 67% of the cases, the pirC homologs are flanked by a gene encoding for a radical S-adenosyl methionine (SAM)-like protein, annotated as Elongator protein 3 (SI Appendix, Fig. S1), which was recently shown to be a noncanonical transfer RNA acetyltransferase (23). In Synechocystis, the pirC gene is upstream of glgA1, which encodes the major glycogen synthase that is required for acclimation to nitrogen deprivation (19).
Protein sequence alignments showed that the first 52 N-terminal amino acids of the annotated PirC sequence are not conserved in any of the other orthologs (SI Appendix, Fig. S2). Furthermore, the experimentally validated transcriptional start site from Synechocystis (24) suggests a shorter open reading frame (ORF) with Met-53 as putative translational start site for PirC. Our initial heterologous expression of the long and short pirC variant in Escherichia coli revealed that only the short variant can be expressed into a properly folded and soluble protein. This finding supports the notion that the 112-amino-acid–long version of PirC represents the physiologically relevant protein. Hence, this short variant was used in all subsequent work.
PirC Is a Strong PII-Binding Partner.
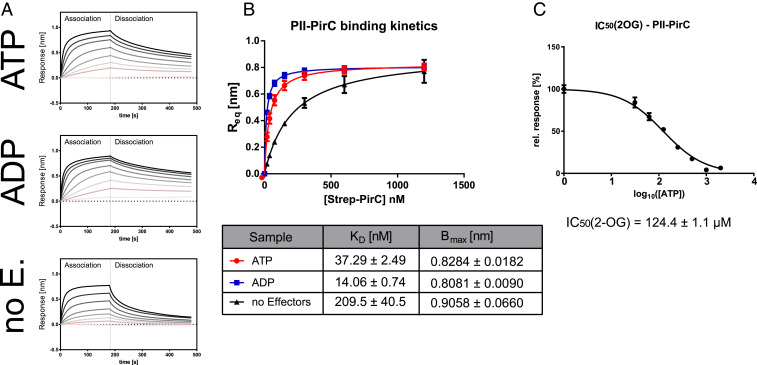
A first series of experiments were set out to verify the putative interaction between PII and PirC (12). In vitro affinity purification experiments revealed that recombinant PirC coeluted with strep-tagged PII in the presence of ATP or ADP, but not in the presence of ATP plus 2-OG (SI Appendix, Fig. S3A). As expected, in the control samples lacking strep-tagged PII, the elution fractions did not contain any PirC. The observation that PirC is unable to interact with ATP- and 2-OG–bound PII is common to many PII-interacting partners and a sign of binding specificity (25). The influence of effector molecules on the interaction between PirC and PII was further quantitatively analyzed by bio-layer interferometry (BLI). In this assay, binding of an analyte in solution to a ligand immobilized on a biosensor surface (or tip) produces a shift in wavelength, which serves as readout of analyte–ligand interaction. For our purposes, recombinant C-terminal His8-tagged PII was bound to the Ni-NTA biosensor surface, while strep-tagged PirC was added as analyte in solution. In the absence of effector molecules, we observed a weak interaction between PII and PirC (Fig. 1 A and B). In the presence of ATP or ADP, PirC binding to PII strongly increased. Quantitative measurements revealed apparent KD values of 37.3 ± 2.5 nM and 14.1 ± 0.7 nM for PII–PirC complex formation in the presence of 2 mM ATP and ADP, respectively. Similar results were obtained using surface plasmon resonance (SPR) spectrometry. The maximum response after injection of PirC in presence of ATP over the PII-loaded sensor was 270 response units (RUs) when the SPR sensor chip was loaded with 1,000 RUs of His-tagged PII (SI Appendix, Fig. S3B). Given that in SPR spectrometry, the response signal in RUs is proportional to the mass change on the sensor, the mass increase of 270 RUs by PirC on 1,000 RUs of PII-loaded sensor is close to one PirC monomer per PII trimer bound. The inhibitory effect of 2-OG on PII–PirC interaction was further quantified by BLI through titration with increasing 2-OG concentrations at a constant concentration of 2 mM ATP (Fig. 1C). A half maximal inhibitory concentration (IC50) of 123.4 ± 1.1 µM for 2-OG was determined, a value close to the KD of the third (lowest affinity) 2-OG–binding site of PII (26). Therefore, it seems that occupation of all three effector binding sites in PII with 2-OG is required to prevent complex formation with PirC.
Fig. 1.
Complex formation of PII with PirC and modulation by effectors ADP, ATP, and 2-OG. (A) BLI binding assays. His-tagged PII was immobilized on sensor tips and allowed to associate for 180 s with PirC in presence of either 2 mM ATP (Top), ADP (Middle), or without effectors (Bottom), followed by 300 s dissociation. The overlay of response curves with increasing concentrations of PirC (9.375 nM to 1,500 nM) is shown. (B) Plot of maximum binding responses from (A) for the calculation of binding constants (depicted below). (C) Plot of IC50 determination for inhibition of PirC–PII binding by increasing 2-OG concentrations at a constant 2 mM of ATP. All experiments were performed in triplicates, and corresponding SDs are shown in B and C.
Physiological Role of PirC in Synechocystis.
The high conservation of pirC in the cyanobacterial phylum, including conservation of the NtcA-binding site, indicated an important function for PirC during acclimation to nitrogen depletion, a feature common to all the members of this phylum. To identify such a function, we generated a pirC-deficient mutant (ΔpirC) as well as strains complemented either with the native pirC gene (ΔpirC::pirC) or with pirC variants encoding fluorescent proteins fused to PirC (SI Appendix, Fig. S4).
Acclimation of these strains to long-term nitrogen starvation was investigated under continuous light or in a day/night regime. Growth (as indicated by an increase in optical density) and degree of pigmentation as well as glycogen and PHB content were monitored over 1 mo. In the wild-type strain pigment degradation after removal of combined nitrogen required 21 d under day/night regimes but only 5 to 7 d in continuous light (SI Appendix, Fig. S5) (15). Pigment degradation was slightly retarded in the ΔpirC mutant compared to the wild-type and complemented strain. Moreover, the increase in optical density at 750 nm (OD750) of the ΔpirC mutant was lower than that of the wild-type and complemented strains (Fig. 2A), which indicates that in the ΔpirC mutant, the final cell division upon nitrogen deprivation is delayed. In contrast to these rather modest effects, a striking phenotype for the ΔpirC strain was observed with respect to glycogen accumulation. After the removal of combined nitrogen sources, both the wild-type and complemented strain showed the typical rapid and steep increase in cellular glycogen levels, which were maintained throughout the entire period of nitrogen-starvation–induced chlorosis. By contrast, glycogen content in the ΔpirC mutant reached only 28% of the wild-type level and subsequently declined again. As opposed to glycogen, the ΔpirC mutant accumulated significantly more PHB (up to 49% of the cell dry mass) than the wild-type and the complemented strain (30% and 29% PHB per cell dry mass, respectively) (Fig. 2B). To confirm this result, PHB granules were visualized by fluorescence microscopy after staining the cells with Nile Red or by transmission electron microscopy (TEM) (Fig. 2C). After 35 d of nitrogen depletion, a much higher PHB content was observed in the ΔpirC mutant than in the wild-type or complemented strain in both fluorescent and TEM micrographs, confirming the results of the chemical PHB quantification.
Fig. 2.
Effect of PirC deletion on growth, carbon storage, and carbon polymer accumulation during chlorosis. The graphs represent the mean and SD from three biological replicates. (A) Growth curves of the wild-type (WT), the pirC null mutant (ΔpirC), and the complemented (ΔpirC::pirC) strain as measured by OD750 starting 7 d before nitrogen depletion and during the 35 d of acclimation to nitrogen starvation, also known as chlorosis. (B) Glycogen and carbon polymer (PHB) content during chlorosis. PHB: plane lines; Glycogen: dashed lines. (C) Fluorescent and TEM micrographs of cells stained with Nile Red after 35 d of chlorosis. Top rows: three-dimensional (3D)–deconvoluted overlay pictures of phase contrast- and fluorescence microscopy images (1,000× magnification). Note that Nile Red stains the PHB granules within each cell. Bottom rows: TEM Pictures (5,000× magnification).
Identification of PirC-Controlled Processes.
The altered glycogen and PHB accumulation patterns in nitrogen-deprived ΔpirC cells suggested a crucial role for PirC in carbon storage metabolism during nitrogen starvation. To elucidate the corresponding molecular mechanism, we aimed to identify additional molecular targets of PirC. To this end, coimmunoprecipitation (CoIP) experiments were conducted using crude extract of nitrogen-starved ΔpirC cells expressing a PirC–mCitrine fusion protein (ΔpirC::pirC–mCitrine). PirC–mCitrine in the crude extract of ΔpirC::pirC–mCitrine was precipitated using a GFP-trap consisting of an anti-GFP Nanobody/VHH coupled to magnetic agarose beads (http://www.chromotek.com). Note that the anti-GFP nanobodies bind different variants of GFP, including mCitrine. Chromotek binding control magnetic agarose was used to determine the unspecific background binding. IPs were performed in the presence of Mg2+, ATP, and 2-OG or in the absence of additionally supplemented effectors. The eluates from independent experiments were analyzed after tryptic digestion by quantitative mass spectrometry to identify coimmunoprecipitating proteins. In the absence of 2-OG, immunoprecipitation of PirC–mCitrine only enriched for PII, confirming that PII is the major PirC-interaction partner in these conditions (SI Appendix, Fig. S6). The addition of 2-OG/ATP to the extract completely changed the pattern of coimmunoprecipated proteins: instead of PII, the enzyme 2,3-bisphosphoglycerate–independent PGAM, encoded by the gene slr1945, appeared as dominant PirC interactor (SI Appendix, Fig. S7). PGAM converts 3-phosphoglycerate (3-PGA) into 2-phosphoglycerate (2-PGA) at the beginning of lower glycolysis. In addition to PGAM, an ortholog of the CcmP protein, encoded by the gene slr0169, was also found as a PirC-interacting protein, but with a lower enrichment factor compared to PGAM. CcmP has been identified as a minor shell protein in carboxysomes of Synechococcus elongatus PCC 7249. The trimeric shell protein has a central pore that can be opened and closed, most likely for the movement of metabolites such as the PGAM substrate 3-PGA (27, 28).
PirC Swaps from PII- to PGAM-Binding in a 2-OG–Dependent Manner, Thus Inhibiting PGAM Activity during Chlorosis.
The observed interaction of PirC with PGAM suggested that PirC negatively regulates PGAM activity. This assumption is consistent with the decreased glycogen and increased PHB levels in the ΔpirC mutant because PGAM diverts newly fixed carbon from the Calvin cycle toward lower glycolysis, through which acetyl-CoA, the precursor metabolite of PHB, is produced. To validate the putative role of PirC in the regulation of PGAM, recombinant PGAM was purified via an N-terminal His6-tag for biochemical characterization.
First, we tested the influence of PirC on PGAM catalytic activity using an enzymatic assay, in which the PGAM-catalyzed conversion of 3-PGA to 2-PGA is coupled with enolase, pyruvate kinase, and lactate dehydrogenase to the final oxidation of reduced nicotinamide adenine dinucleotide (NADH). The His-tag was removed from recombinant PGAM by thrombin cleavage to prevent interference with catalysis. Furthermore, we verified that PirC had no effect on the activities of the coupling enzymes (SI Appendix, Fig. S8). A clear PirC-dependent inhibition of PGAM activity was observed when PirC was added at increasing concentrations to the reaction mix of the enzymatic assay. PirC inhibited PGAM in a competitive manner by increasing the KM for the substrate 3-PGA rather than lowering the Vmax. At nearly equimolar concentrations of PirC (200 nM) and PGAM (166 nM), the catalytic efficiency was reduced to one third. In the presence of an excess of PirC, the catalytic activity of PGAM could be reduced more than 10-fold as compared to the absence of PirC (Fig. 3A and Table 1).
Fig. 3.
Effect of PirC on PGAM enzyme activity and PGAM–PirC complex formation. (A) Inhibition of PGAM activity by increasing PirC concentrations represented as Lineweaver–Burk plot. The corresponding kinetic constants are shown in Table 1. PirC has no effect on coupling enzymes as shown in supplements (SI Appendix, Fig. S8). (B) Modulation of PGAM activity by PirC (600 nM) in presence or absence of PII (600 nM trimer) and 0.4 mM ATP without or with 0.123 mM or 1 mM 2-OG. Each point represents the mean of triplicates. (C) Steady-state graph of PirC–PGAM binding assays using BLI. The mean of the Req value (three independent replicates) was plotted against the molar concentration of Strep–PirC. The inset shows the raw binding curves at different PirC concentrations. (D) Competitive inhibition of PirC–PGAM interaction by 2-PGA and 3-PGA. Plot shows determination of IC50. (E) Steady-state graph of PirC–PGAM binding in presence of 2-PGA or 3-PGA at their IC50 concentrations.
Table 1.
Kinetic constants of PGAM under varying concentrations of PirC and changes of constants by addition of PirC-interacting molecules
| Inhibition of PGAM by PirC at varying concentrations | ||||
| PirC, nM | KM, mM | vmax, nkat · mg−1 | kcat, s−1 | kcat · KM−1, s−1 · M−1 |
| 0 | 0.1266 ± 0.0064 | 47.54 ± 0.52 | 2.776 ± 0.030 | 21959.0 ± 1104.7 |
| 200 | 0.3584 ± 0.0216 | 44.92 ± 0.75 | 2.623 ± 0.044 | 7329.8 ± 441.1 |
| 400 | 0.7056 ± 0.0499 | 46.56 ± 1.09 | 2.719 ± 0.063 | 3859.1 ± 273.0 |
| 600 | 1.376 ± 0.102 | 47.11 ± 1.39 | 2.751 ± 0.081 | 2002.2 ± 148.4 |
| 800 | 1.715 ± 0.145 | 43.27 ± 4.10 | 2.526 ± 0.239 | 1618.7 ± 369.4 |
| Antagonistic effect of PII and its modulation by 2-OG (0.4 mM ATP, 600 nM PirC, and 600 nM PII3) | ||||
| Modulators of PGAM | KM, mM | vmax, nkat · mg−1 | kcat, s−1 | kcat · KM−1, s−1 · M−1 |
| None | 0.1499 ± 0.0121 | 89.76 ± 2.17 | 5.41 ± 0.13 | 36097.4 ± 2921.0 |
| PirC | 4.0220 ± 1.338 | 92.80 ± 24.78 | 5.59 ± 1.49 | 1390.9 ± 462.7 |
| PirC/PII | 0.1855 ± 0.0247 | 105.90 ± 4.51 | 6.38 ± 0.27 | 34409.7 ± 4572.5 |
| PirC/PII + 0.123 mM 2-OG | 1.8410 ± 0.5461 | 142.70 ± 28.47 | 5.56 ± 1.24 | 3021.7 ± 896.3 |
| PirC/PII + 1 mM 2-OG | 2.887 ± 0.8542 | 92.28 ± 20.51 | 8.60 ± 1.72 | 2980.3 ± 881.8 |
| Binding constants for PGAM–PirC binding and modulation by substrates | ||||
| Sample | KD, nM | IC50, mM | ||
| No Effectors | 2.89 ± 34.49 | — | ||
| 2-PGA | 1702 ± 744 (at IC50) | 0.97 ± 0.001 | ||
| 3-PGA | 459.1 ± 32.0 (at IC50) | 2.4 ± 0.001 | ||
Binding constants of PGAM/PirC complex with or without the presence of the substrates.
Second, the interplay of PII and PirC on PGAM activity was analyzed because we assumed that PII might regulate the inhibitory interaction of PirC with PGAM, in analogy to the effect of PII on PipX–NtcA interaction (14). To this end, we performed PGAM assays in the presence of PirC and PII and supplemented the assays with ATP and different 2-OG concentrations, respectively (Fig. 3B). Addition of PII in the absence of 2-OG abolished the inhibitory effect of PirC on PGAM activity. The KM for 3-PGA returned to the value of noninhibited PGAM (Table 1). However, in the presence of 1 mM of 2-OG, a concentration corresponding to high C/N conditions in Synechocystis cells, PirC was again able to inhibit PGAM as in the absence of PII. When 2-OG was added at a concentration of 0.123 mM (corresponding to the IC502-OG value of PII–PirC complex formation), the inhibition of PGAM was ∼50% of the maximal inhibition with PirC in the absence of 2-OG. These results unambiguously indicate that in vitro, in presence of high 2-OG levels, binding of 2-OG to PII disrupts PII–PirC interaction and promotes binding of PirC to PGAM, thus inhibiting its activity.
The inhibitory effect of PirC binding on PGAM activity was further investigated using BLI assays. To this end, His6-tagged PGAM was bound to the biosensor tip, and strep-tagged PirC protein was added in solution as analyte at varying concentrations (Fig. 3C). A stable PGAM–PirC complex formed in the absence of any effector molecules. The competitive inhibition mode (Fig. 3A) suggested that the substrates of PGAM could compete with PGAM–PirC interaction. Indeed, the addition of the PGAM substrates 3-PGA or 2-PGA (for the forward and backward reaction, respectively) had inhibitory effects on complex formation. Addition of 2-PGA inhibited complex formation 2.4 times stronger (IC50 = 0.97 mM) than 3-PGA (IC50 = 2.4 mM) (Fig. 3D). When the metabolites were added at their IC50 concentration, 2-PGA (1 mM) increased the KD of PirC–PGAM interaction to 1,702 nM and 3-PGA (2.4 mM) to 459 nM, respectively (Fig. 3E and Table 1).
PirC Deletion Leads to Accumulation of the Metabolites of Lower Glycolysis.
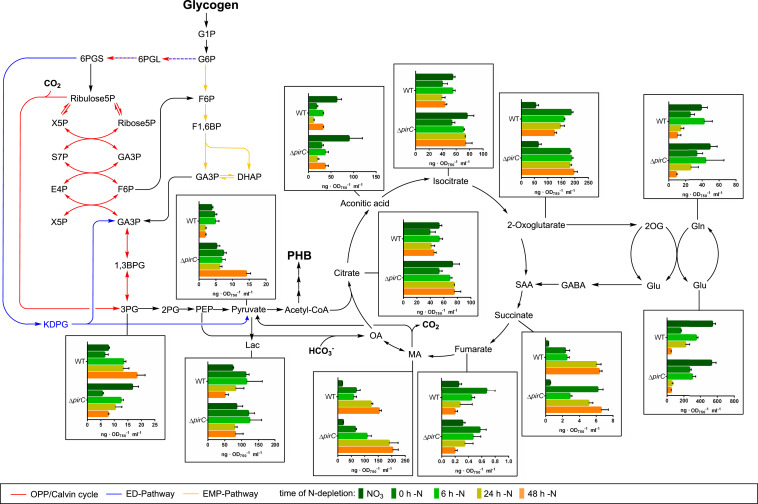
The above-described analysis of the PGAM–PirC–PII triad demonstrated inhibition of PGAM activity by PirC, in response to the binding of the C/N-status reporter metabolite 2-OG to PII. This suggests that in wild-type cells, during nitrogen starvation (i.e., when high 2-OG levels accumulate), the inhibition of PGAM by PirC supports the formation of high glycogen levels by diminishing carbon catabolism via lower glycolysis. In the absence of this inhibition (i.e., in the ΔpirC mutant), glycogen catabolism via glycolysis increases. To further verify this hypothesis, we monitored over time the levels of metabolites in wild-type and ΔpirC mutant cells during the shift from nitrate-replete (NO3) to nitrogen-depleted (−N) medium. Samples for metabolome analysis were withdrawn after 0, 6, 24, and 48 h from the shift. Nitrogen depletion had the expected effect on the total cellular steady-state metabolite pools (i.e., soluble amino acids were depleted to large extent, while organic acids accumulated), resulting in lowered N/C ratios under −N conditions (SI Appendix, Fig. S9). Most organic acids participating in the TCA cycle such as citrate, malate, and succinate accumulated in both strains in a similar manner when shifted to –N conditions (Fig. 4). Also, the products of ammonium assimilation via GS/GOGAT, glutamine (Gln), and glutamate (Glu) showed similar changes in the wild-type and ΔpirC strain with rapid decrease in Glu and slower decrease in Gln. Besides these general metabolic responses, in which the ΔpirC mutant showed no discernable differences, a few very specific and intriguing differences were recorded at decisive steps: the C/N-status reporter molecule 2-OG accumulated immediately after −N shift in both strains. In the wild-type, the 2-OG levels decreased gradually over the following 48 h, whereas they remained constantly elevated in ΔpirC cells, and they even slightly increased (Fig. 4). Moreover, the 3-PGA concentration increased in the wild-type over the course of nitrogen starvation, whereas it gradually declined in the ΔpirC mutant. The increasing 3-PGA levels, substrate of the PGAM, in the wild-type cells indicate in vivo inhibition of the PGAM reaction, whereas lack of PGAM inhibition by PirC explains the low 3-PGA levels in the ΔpirC mutant. 3-PGA is known as an allosteric activator of the glucose 1-phosphate–adenylyltransferase (GlgC) in bacteria, which catalyzes the initial step of glycogen synthesis (29). Downstream of the PGAM reaction, the levels of pyruvate responded inversely to 3-PGA, with decreasing levels in the wild-type but a strong increase in the ΔpirC mutant. The pyruvate level in the mutant was 14-fold higher than in the wild-type after 48 h of N starvation. Again, this observation is consistent with an increased flux through the PGAM reaction due to the missing inhibition by PirC, since the produced 2-PGA is further converted into pyruvate. The increased carbon flux toward pyruvate in the ΔpirC mutant lowers the carbon flux toward glycogen and increases the levels of PHB, which is derived from acetyl-CoA (i.e., the immediate reaction product from pyruvate).
Fig. 4.
Time course LC-MS analysis of steady-state levels of relevant metabolites during the short-term shift of wild-type (WT) and ΔpirC cells from NO3 to −N conditions depicted in a metabolic background. Totals of 0 h, 6 h, 24 h, and 48 h indicate the time from nitrogen depletion at which samples were withdrawn from the cultures for analysis. Each bar represents the metabolite level at a certain time point determined from two independent biological replicates each in technical duplicates. The error bars represent the SD of the combined data. The values are in ng per optical density at 750 nm per ml (ng · OD750−1 · ml−1). The result of the entire metabolite analysis is shown in SI Appendix, Fig. S9.
Subcellular Localization of PirC.
To determine the subcellular localization of PirC during different growth stages, when PirC presumably interacts preferentially with either PII or PGAM, we analyzed cells expressing a PirC–GFP fusion protein (ΔpirC::pirC–eGFP) by fluorescence microscopy. The eGFP signal was centrally localized in the cytoplasm (SI Appendix, Fig. S10) in cells during exponential growth in nitrate-containing BG11 medium, conditions that promote binding of PirC to PII. Shifting the cells to nitrogen-depleted medium increased the 2-OG levels (see Fig. 4), which should promote dissociation of the PII–PirC complexes and allow interaction of PirC with PGAM. Accordingly, the localization of PirC–eGFP changed after nitrogen downshift. The centrally localized eGFP signal slowly expanded to the peripheral region of the cytoplasm during the first 24 h after nitrogen starvation, where it then remained throughout chlorosis. This result corroborated the dynamics of PirC interactions and its response to nitrogen limitation.
Discussion
In this study, we identified a key control point of cyanobacterial carbon metabolism, the glycolytic PGAM reaction, converting 3-PGA into 2-PGA. In animal systems, where glycolysis supplies energy, it has been shown that glycolytic breakdown of glucose 6-phosphate is mainly regulated at the phosphofructokinase level according to the energy demand of the cells (30). By contrast, in photoautotrophic organisms, glycolytic steps are used in two directions, in the gluconeogenic direction toward glycogen or starch synthesis and in the glucose catabolic direction, respectively, to produce precursors for multiple biosynthetic routes required for cell growth. The PGAM reaction is at the branch point of newly fixed CO2. 3-PGA, the first stable reaction product from RubisCO-catalyzed CO2 fixation, can be metabolized in two directions. Most of it is converted into 2,3-bisphosphoglycerate and further to glyceraldehyde-3-phosphate (GAP), from which the acceptor of RubisCO, ribulose 1,5-bisphosphate, is regenerated via the Calvin cycle reactions. Excess GAP is used via gluconeogenic reactions to synthesize glycogen (in plant starch). Alternatively, 3-PGA can be diverted from the Calvin cycle through its direct conversion to 2-PGA by PGAM. 2-PGA is further metabolized in lower glycolytic reactions, from where the majority of cellular amino acids and lipids are derived in photoautotrophs, with pyruvate, acetyl-CoA, and 2-OG representing key precursors.
The PGAM reaction was previously predicted as a key control point of carbon metabolism by kinetic modeling of the cyanobacterial low-carbon response (20, 21). It was shown that 2-PGA accumulates to high amounts (5 to 7 times) in cells shifted from high CO2 (5%) to ambient air (0.04% CO2) in Synechocystis (31) as well as in Synechococcus elongatus PCC 7942 (32). The high 2-PGA accumulation was taken as an indication that under carbon-limiting conditions, newly fixed organic carbon is directly deviated from the Calvin cycle into lower glycolysis by the PGAM reaction to sustain biosynthesis of amino acids and other cellular compounds. Here, we provide in vitro and in vivo evidence that the reaction catalyzed by the product of the slr1945 gene, PGAM, represents a key control point for acclimation to nitrogen starvation. This control operates through a regulatory mechanism, in which the small regulatory protein PirC acts as a mediator of the signal from the pervasive PII regulatory protein to tune the activity of PGAM, a control mechanism so far never described for enzymatic reactions. To further understand the competitive inhibition of PGAM by PirC, as demonstrated here through kinetic and binding studies, structural analysis of the enzyme complexes will be required.
According to our model depicted in Fig. 5, PII binds to PirC under nitrogen-sufficient conditions, when 2-OG levels are low, thereby preventing the interaction with PGAM. Efficient conversion of 3-PGA to 2-PGA by highly active PGAM directs newly fixed carbon toward lower glycolysis to support the synthesis of amino acids and fatty acids. Only a minor fraction is converted into glycogen. When the cells experience nitrogen limitation, they accumulate the intracellular 2-OG levels. As a result of 2-OG binding to PII, the PII–PirC complex dissociates, and PirC interacts with PGAM, thereby inhibiting its enzymatic activity. This is accompanied by a relocalization of the PirC–eGFP signal from the central region of the cell to the periphery. The central localization of PirC–eGFP is indicative of the presence of PirC–PII complexes as PII was previously shown to localize in the central cytoplasm of cells grown in the presence of nitrate (11). As a consequence of the PirC–PGAM interaction, conversion of 3-PGA to 2-PGA is blocked in nitrogen-starved cells, leading to increased 3-PGA levels, which are now redirected toward glycogen. Due to this metabolic switch, the flux toward amino acid synthesis is slowed down, thus adjusting cellular metabolism to the limited supply of nitrogen. Furthermore, 3-PGA is an allosteric activator of GlgC, which catalyzes the initial and regulated step of the glycogen synthesis (29). Hence, the PirC-mediated PGAM inhibition not only slows down lower glycolysis but also stimulates glycogen accumulation via the GlgC activation. The glycogen levels increase until the cells are densely packed with glycogen, which can amount up to 50% of the dry weight in chlorotic cells (15). Already after 24 h of nitrogen starvation, accumulation of glycogen reaches a maximum, and the levels remain high throughout chlorosis. Recent data indicate a constant turnover of glycogen until the cells enter complete dormancy (17). In Synechocystis, which expresses the PHB synthesis machinery, acetyl-CoA molecules arising from the residual glycolytic flux during chlorosis are directed toward PHB synthesis. Hence, the amount of PHB steadily increases during prolonged nitrogen starvation. In the ΔpirC mutant, PGAM cannot be appropriately inhibited. Therefore, increased flux toward 2-PGA and lower glycolysis leads to a massive accumulation of PHB. In agreement with this model, PII-deficient Synechocystis mutants are unable to accumulate PHB during nitrogen-starvation–induced chlorosis (33). In the absence of PII, PirC will constantly inhibit PGAM activity, resulting in decreased levels of metabolites downstream of 2-PGA (10) due to the limited supply of acetyl-CoA for PHB synthesis.
Fig. 5.
Model of regulation of central carbon metabolism by PirC, PII, and PGAM interactions. In vegetative cells, when 2-OG levels are low, PII (depicted as trimer in blue, green, and red) binds to PirC and prevents the inhibition of PGAM. PGAM directs 3-PGA downstream to the biosynthesis of fatty acids (FAS) and amino acids mainly via the TCA cycle. When the cells are N depleted, 2-OG levels increase and promote release of PirC from PII. PirC binds to and inhibits PGAM. This results in an elevation of the 3-PGA concentration. 3-PGA enhances the activity of GlgC and directs the carbon flux toward glycogen, resulting in enhanced glycogen accumulation. CBB: Calvin-Benson-Bassham cycle.
This glycolytic switch at the enzymatic level of PGAM via the small PII-interacting regulatory protein PirC is reminiscent of the control of NtcA-dependent transcription by the small PII-interacting protein PipX. The latter is either complexed to PII under low 2-OG levels or bound to NtcA at elevated 2-OG levels (6). Hence, the two small PII-mediator proteins, PirC as well as PipX, functionally interact to coherently reprogram metabolism and gene expression under low-nitrogen conditions. Through release of the PII–PirC complex in response to increasing 2-OG levels, PirC tunes down PGAM activity. This response is further amplified by the concomitant dissociation of the PII–PipX complex and association of PipX to NtcA, accompanied by NtcA-dependent expression of many low-nitrogen–induced genes, among which is the pirC gene. In chlorotic cells, pirC (sll0944) is among the most strongly up-regulated genes within the entire transcriptome (15), and, accordingly, PirC is one of the most highly enriched proteins during chlorosis (16). In an advanced stage of chlorosis, this strong accumulation of PirC ensures tight inhibition of glycolysis to maintain high glycogen levels, which are required for efficient exit from the dormant state of chlorosis (19).
Through engagement of such mediator proteins, PII can largely expand its regulatory space. In principle, any cellular activity, which can be modulated through interaction with a peptide, could be tuned by PII by engineering the peptide to bind PII, thus providing a new toolbox for synthetic biology. It is also reasonable to speculate that disabling the regulation of PirC can be used for metabolic engineering in cyanobacteria, in particular, for the bioproduction of metabolites derived from lower glycolysis, such as succinate, malate, or lipids as well as fatty acids. Furthermore, cyanobacteria have the potential to synthesize isoprenoids or terpenes via the methylerythritol phosphate pathway whose precursors are pyruvate and GAP (34). In addition, genetically engineered cyanobacteria can be used for the synthesis of heterologous compounds such as l-butanol from acetyl-CoA (35). However, it should be kept in mind that the utility of the PirC mutation is especially beneficial for processes that are performed under nitrogen-limited conditions. As a proof of principle, we took advantage of the redirection of the carbon metabolism toward PHB synthesis in the PirC-deficient mutant to further increase the levels of PHB in the cell: we engineered the PirC-deficient mutant to express the high processive PHB biosynthetic enzymes from Cupriavidus necator, resulting in a strain that accumulates more than 80% PHB of cell dry mass under nitrogen-depleted conditions. This is by far the highest accumulation of PHB ever reported in a cyanobacterium (36). This result impressively demonstrates the impact and biotechnological potential of decoupling the regulation by PirC.
Materials and Methods
Full protocols are available in SI Appendix, SI Materials and Methods.
Strains and Cultivation.
A list of all used strains for this study is provided in SI Appendix, Table S1. Synechocystis sp. PCC 6803 strains were cultivated in BG11 medium according to Rippka (37) either at continuous illumination (∼50 μE m–2 · s–1) or light–dark conditions (12 h light and 12 h darkness) and 28 °C. For nitrogen depletion, cultures were cultivated in BG-11 media without 17.65 mM NaNO3. Whenever necessary, appropriate antibiotics were added to the different strains to ensure the continuity of the mutation. Cultivation of E. coli cultures was performed with lysogeny broth (LB) medium and LB-agar.
Plasmids and Cloning.
A list of all used primers, plasmids, and the cloning procedures is in SI Appendix, Table S2 as well as in SI Appendix, Fig. S4.
Overexpression and Purification of Proteins.
Recombinant proteins were overexpressed in E. coli Lemo21. His-tag proteins were purified on 1 mL Ni-NTA HisTrap columns (GE Healthcare) as described previously (10). For purification of strep-tagged proteins, 5 mL strep-tactin superflow columns were used as described previously (10). The His-tag of PGAM was removed by Thrombin cleavage using bovine thrombin of Sigma-Aldrich according to protocol (38).
CoIP and Liquid Chromatography–Mass Spectrometry.
For CoIP experiments in presence of the PII effector molecules, cells of Synechocystis ΔPirC::PirC–mCitrine cultures were harvested after 24 h N depletion and resuspended in 2 mL binding buffer (100 mM Tris [pH 7.5], 100 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.5 mM EDTA, 2 mM ATP, and 2-OG). After lysing the cells, the lysate was centrifuged, and from the supernatant, a volume containing 3 mg of protein was used for the immunoprecipitation using GFP-Trap Magnetic Agarose beads or control beads without antibodies according to the manufacturer’s protocol (Chromotek, Planegg-Martinsried, Germany). Bound proteins were eluted by heating in sodium dodecyl sulfate (SDS) loading buffer, and the solution was subjected to short SDS-polyacrylamide gel electrophoresis (PAGE) runs on 12% Bis-Tris Gels (Invitrogen). After staining with Coomassie blue, protein regions were isolated and InGel digested with trypsin as described (39). After cleaning peptides with StageTips (40), liquid chromotography–mass spectometry (LC-MS/MS) analysis was performed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, Germany), using linear, segmented 60-min nano liquid chromatography (nanoLC) reversed phase (RP) gradients as described (16). From triplicate experiments, all raw data were processed using MaxQuant software suite (version 1.6.5.0) at default settings. Tandem mass spectrometry peak lists were searched against a target-decoy database of the Synechocystis proteome, including the sequence of PirC (Sll0944)–mCitrine. Label-free quantification (LFQ) was used to calculate LFQ intensities for each CoIP sample as described in extended protocol in the supplement.
BLI Using the Octet K2 System.
In vitro binding studies were done by BLI using Octet K2 system (FortéBio) as described previously (10). In the first step, PII-His8 (400 nM, trimeric) or PGAM-His6 (500 nM) was immobilized on Ni-NTA sensors (FortéBio), followed by a 60-s baseline measurement. For the binding of PirC, the biosensors were dipped into the PirC solution for 180 s (association), with concentrations ranging between 9.375 nM to 1,500 nM. In PII-binding studies, effector molecules ADP, ATP, or 2-OG were added to the binding buffer as indicated and in PGAM binding assays, 2PG or 3 PGA. Finally, the complexes were allowed to dissociate for 300 s. For each binding experiment, a negative control without an interaction partner was performed in parallel. The response in equilibrium (Req) was calculated using the data analysis software of the Octet System. To calculate the dissociation constant KD, the concentration versus Req plots were made for each set of experiments.
Glycogen Measurement and PHB Quantification.
Glycogen was quantified according to ref. 19. PHB was detected by high-performance LC as described previously (41).
PGAM Enzymatic Assay.
The PGAM activity and the effect of PirC was determined by a coupled enzyme assay as described previous (42, 43) with 10 µg of purified PGAM used in a 1 mL reaction. The reaction mixture containing 20 mM Hepes-KOH (pH 8.0), 100 mM KCl, 5 mM MgSO4, 0.4 mM MnCl2, 50 µg·ml−1 BSA, 1 mM DTT, 0.4 mM ADP, 0.2 mM NADH, 0.5 U enolase (Sigma Aldrich), 2 U Pyruvate kinase (Sigma Aldrich), 2 U Lactate dehydrogenase (Roche), and 10 µg PGAM was prewarmed to 30 °C. The assay was started by adding the 3-PGA solutions. The resulted decrease of NADH over time was recorded with Specord50 (Jena Analytics) at 340 nm. As blank, an assay without 3-PGA was performed.
Fluorescence Microscopy and TEM.
The visualization of PHB granules was done by phase contrast fluorescence microscopy using the Leica DM5500 B with the Leica CTR 5500 illuminator as described previously. Electron microscopic pictures of lead-citrate– and uranyl-acetate–stained microtome sections of glutaraldehyde and potassium-permanganate–fixed Synechocystis cells were prepared as described (44). The samples were then examined using a Philips Tecnai 10 electron microscope at 80 kHz.
Metabolome Analysis.
For metabolome analysis by LC-MS, Synechocystis was cultivated in 200 mL under N depletion as described previously for 48 h under continuous lightning. The sampling was carried out 0, 6, 24, and 48 h after the shift. Samples of 5 mL liquid culture were quickly harvested onto nitrocellulose membrane filters and subjected to metabolome analytics as detailed in SI Appendix.
Supplementary Material
Acknowledgments
The project was funded by grants from the German Research Foundation (DFG) Fo195/9-2 and as part of the research group (FOR2816) SCyCode “The Autotrophy-Heterotrophy Switch in Cyanobacteria: Coherent Decision-Making at Multiple Regulatory Layers” to M.H. (HA 2002/23-1) and B.M. (MA 4918/4-1). LC-MS/MS systems at the Department of Quantitative Proteomics were supported through the DFG grants INST 37/935-1 and INST 37/741-1 FUGG. The LC-MS/MS equipment at University of Rostock was financed through the HBFG program (GZ: INST 264/125-1 FUGG). The work was further supported by infrastructural funding from the DFG Cluster of Excellence EXC 2124 Controlling Microbes to Fight Infections. We thank Libera Lo Presti for proofreading the manuscript, Karl-Heinz Grenzendorf for technical assistance in protein purification, and Claudia Menzel for TEM preparations. We want to acknowledge Alexander Klotz for initial contributions on Sll0944 research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2019988118/-/DCSupplemental.
Data Availability.
Proteome raw data files acquired by mass spectrometry were deposited at the ProteomeXchange Consortium via the Proteomics Identifications Database partner repository (45) under the identifier PXD021415.
References
- 1.Galperin M. Y., A census of membrane-bound and intracellular signal transduction proteins in bacteria: Bacterial IQ, extroverts and introverts. BMC Microbiol. 5, 35 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selim K. A., Ermilova E., Forchhammer K., From cyanobacteria to archaeplastida: New evolutionary insights into PII signalling in the plant kingdom. New Phytol. 227, 722–731 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Forchhammer K., Selim K. A., Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 44, 33–53 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huergo L. F., Dixon R., The emergence of 2-oxoglutarate as a master regulator metabolite. Microbiol. Mol. Biol. Rev. 79, 419–435 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forchhammer K., Lüddecke J., Sensory properties of the PII signalling protein family. FEBS J. 283, 425–437 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Forcada-Nadal A., Llácer J. L., Contreras A., Marco-Marín C., Rubio V., The PII-NAGK-PipX-NtcA regulatory axis of cyanobacteria: A tale of changing partners, allosteric effectors and non-covalent interactions. Front. Mol. Biosci. 5, 91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burillo S., Luque I., Fuentes I., Contreras A., Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J. Bacteriol. 186, 3346–3354 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinrich A., Maheswaran M., Ruppert U., Forchhammer K., The Synechococcus elongatus P signal transduction protein controls arginine synthesis by complex formation with N-acetyl-L-glutamate kinase. Mol. Microbiol. 52, 1303–1314 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y.-M., White S. W., Rock C. O., Inhibiting bacterial fatty acid synthesis. J. Biol. Chem. 281, 17541–17544 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Scholl J., Dengler L., Bader L., Forchhammer K., Phosphoenolpyruvate carboxylase from the cyanobacterium Synechocystis sp. PCC 6803 is under global metabolic control by PII signaling. Mol. Microbiol. 114, 292–307 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Santos A. R. S., et al., NAD+ biosynthesis in bacteria is controlled by global carbon/nitrogen levels via PII signaling. J. Biol. Chem. 295, 6165–6176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watzer B., et al., The signal transduction protein PII controls ammonium, nitrate and urea uptake in cyanobacteria. Front. Microbiol. 10, 1428 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giner-Lamia J., et al., Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 45, 11800–11820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espinosa J., Forchhammer K., Burillo S., Contreras A., Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 61, 457–469 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Klotz A., et al., Awakening of a dormant cyanobacterium from nitrogen chlorosis reveals a genetically determined program. Curr. Biol. 26, 2862–2872 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Spät P., Klotz A., Rexroth S., Maček B., Forchhammer K., Chlorosis as a developmental program in cyanobacteria: The proteomic fundament for survival and awakening. Mol. Cell. Proteomics 17, 1650–1669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch M., Doello S., Gutekunst K., Forchhammer K., PHB is produced from glycogen turn-over during nitrogen starvation in Synechocystis sp. PCC 6803. Int. J. Mol. Sci. 20, 20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch M., Berendzen K. W., Forchhammer A. K., On the role and production of polyhydroxybutyrate (PHB) in the cyanobacterium Synechocystis sp. PCC 6803. Life (Basel) 10, 10 (2020). [Google Scholar]
- 19.Doello S., Klotz A., Makowka A., Gutekunst K., Forchhammer K., A specific glycogen mobilization strategy enables rapid awakening of dormant cyanobacteria from chlorosis. Plant Physiol. 177, 594–603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jablonsky J., Hagemann M., Schwarz D., Wolkenhauer O., Phosphoglycerate mutases function as reverse regulated isoenzymes in Synechococcus elongatus PCC 7942. PLoS One 8, e58281 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jablonsky J., Schwarz D., Hagemann M., Multi-level kinetic model explaining diverse roles of isozymes in prokaryotes. PLoS One 9, e105292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vázquez-Bermúdez M. F., Flores E., Herrero A., Analysis of binding sites for the nitrogen-control transcription factor NtcA in the promoters of Synechococcus nitrogen-regulated genes. Biochim. Biophys. Acta 1578, 95–98 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Lin T.-Y., et al., The Elongator subunit Elp3 is a non-canonical tRNA acetyltransferase. Nat. Commun. 10, 625 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitschke J., et al., An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U.S.A. 108, 2124–2129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeth K., Fokina O., Forchhammer K., Structural basis and target-specific modulation of ADP sensing by the Synechococcus elongatus PII signaling protein. J. Biol. Chem. 289, 8960–8972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fokina O., Chellamuthu V.-R., Forchhammer K., Zeth K., Mechanism of 2-oxoglutarate signaling by the Synechococcus elongatus PII signal transduction protein. Proc. Natl. Acad. Sci. U.S.A. 107, 19760–19765 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai F., et al., The structure of CcmP, a tandem bacterial microcompartment domain protein from the β-carboxysome, forms a subcompartment within a microcompartment. J. Biol. Chem. 288, 16055–16063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larsson A. M., Hasse D., Valegård K., Andersson I., Crystal structures of β-carboxysome shell protein CcmP: Ligand binding correlates with the closed or open central pore. J. Exp. Bot. 68, 3857–3867 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preiss J., Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38, 419–458 (1984). [DOI] [PubMed] [Google Scholar]
- 30.Tanner L. B., et al., Four key steps control glycolytic flux in mammalian cells. Cell Syst. 7, 49–62.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenhut M., et al., Metabolome phenotyping of inorganic carbon limitation in cells of the wild type and photorespiratory mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol. 148, 2109–2120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarz D., et al., Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol. 155, 1640–1655 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hauf W., "Regulation of carbon polymer accumulation in Synechocystis sp. PCC 6803," PhD thesis, University of Tübingen, Tübingen, Germany (2016).
- 34.Pattanaik B., Lindberg P., Terpenoids and their biosynthesis in cyanobacteria. Life (Basel) 5, 269–293 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anfelt J., et al., Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Fact. 14, 167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch M.et al., Maximizing PHB content in Synechocystis sp. PCC 6803: Development of a new photosynthetic overproduction strain. Microb. Cell Fact. 19, 231, 10.1101/2020.10.22.350660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanier R. Y., Deruelles J., Rippka R., Herdman M., Waterbury J. B., Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111, 1–61 (1979). [Google Scholar]
- 38.Guan K. L., Dixon J. E., Eukaryotic proteins expressed in Escherichia coli: An improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192, 262–267 (1991). [DOI] [PubMed] [Google Scholar]
- 39.Shevchenko A., Tomas H., Havlis J., Olsen J. V., Mann M., In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber J., Mann M., Ishihama Y., Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Koch M., Orthwein T., Alford J. T., Forchhammer K., The Slr0058 protein from Synechocystis sp. PCC 6803 is a novel regulatory protein involved in PHB granule formation. Front. Microbiol. 11, 809 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chander M., Setlow B., Setlow P., The enzymatic activity of phosphoglycerate mutase from gram-positive endospore-forming bacteria requires Mn2+ and is pH sensitive. Can. J. Microbiol. 44, 759–767 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Kuhn N. J., Setlow B., Setlow P., Manganese(II) activation of 3-phosphoglycerate mutase of Bacillus megaterium: pH-sensitive interconversion of active and inactive forms. Arch. Biochem. Biophys. 306, 342–349 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Fiedler G., Arnold M., Hannus S., Maldener I., The DevBCA exporter is essential for envelope formation in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 27, 1193–1202 (1998). [DOI] [PubMed] [Google Scholar]
- 45.Perez-Riverol Y., et al., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteome raw data files acquired by mass spectrometry were deposited at the ProteomeXchange Consortium via the Proteomics Identifications Database partner repository (45) under the identifier PXD021415.