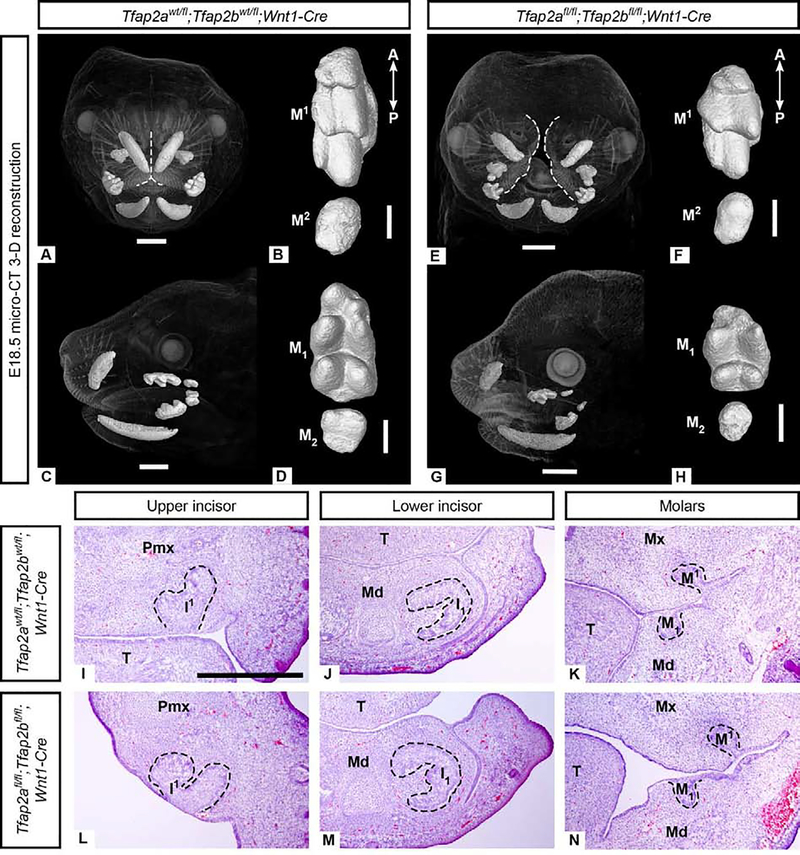
Figure 4. Incisors and molars in Tfap2afl/fl;Tfap2bfl/fl;Wnt1-Cre embryos lacking Tfap2a and Tfap2b expression in the NC-derived mesenchyme lack major morphological defects based on μCT (A-H) and histological (I-N) analyses.
3-D reconstructions of μCT data comparing morphology of control upper (B: M1−2) and lower (D: M1–2) molars with mutant upper (F: M1−2) and lower (H: M1–2) molars. The correct number of cusps are present but the molars appear shorter along the anterior-posterior (A-P) axis than the controls. Note the midface cleft in the mutant (E), outlined in white, compared to the control (A). H&E stained cryosections in the frontal plane (I-N) showing that mesenchyme-specific mutant incisors (L, M) and molars (N) are similar to those of the control (I-K) at the cap stage (E14.5). Due to the cleft midface and palate, in the anterior-most frontal section shown here (L) the medial aspect of the premaxilla is at an angle. Scale bars are 1mm (A, C, E, G) and 300um (B, D, F, H), 500um (I-N). Pmx: premaxilla, Mx: maxilla, Md: mandible, T: tongue.