Fig. 1.
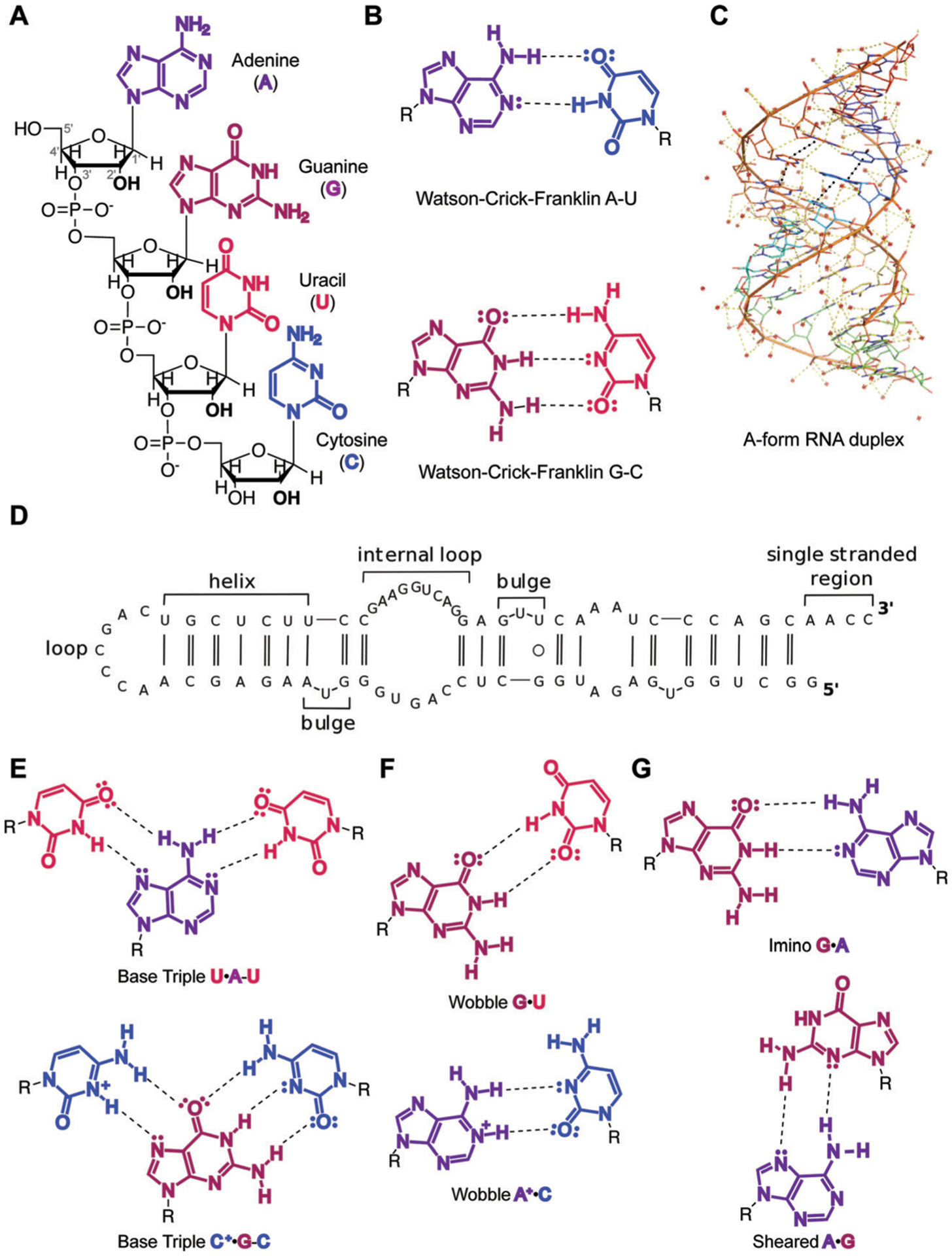
Overview of RNA primary and secondary structure and base pairing patterns. (A) Nucleotide building blocks of RNA primary structure. Each consists of a ribose sugar, containing a 2′ hydroxyl unique to RNA over DNA; phosphodiester groups, which connect the 5′ and 3′ hydroxyl groups of the ribose sugars; and adenine, guanine, uracil and cytosine nucleobases, which define the sequence of an RNA molecule. (B) Canonical Watson–Crick–Franklin hydrogen bonding patterns. Dashed lines represent hydrogen bonds. (C) An A-form RNA duplex secondary structure (PDB ID: 1LNT). Red dots represent metal ions. Yellow dashes indicate electrostatic interactions, including hydrogen bonds. Black dashes indicate stacking interactions. (D) Example of canonical RNA secondary structures as observed in 5′-end of long non-coding RNA (lncRNA) B2-SINE. Single and double solid lines represent A–U and G–C Watson–Crick–Franklin base pairs, respectively, while circles represent wobble base pairs. Adapted from ref. 15 with permission from Elsevier B. V., Copyright 2015. (E) Higher-order base triple interactions. (F) Non-canonical wobble interactions. (G) Purine–purine interactions, including imino and sheared hydrogen bonding patterns.
