Fig. 2.
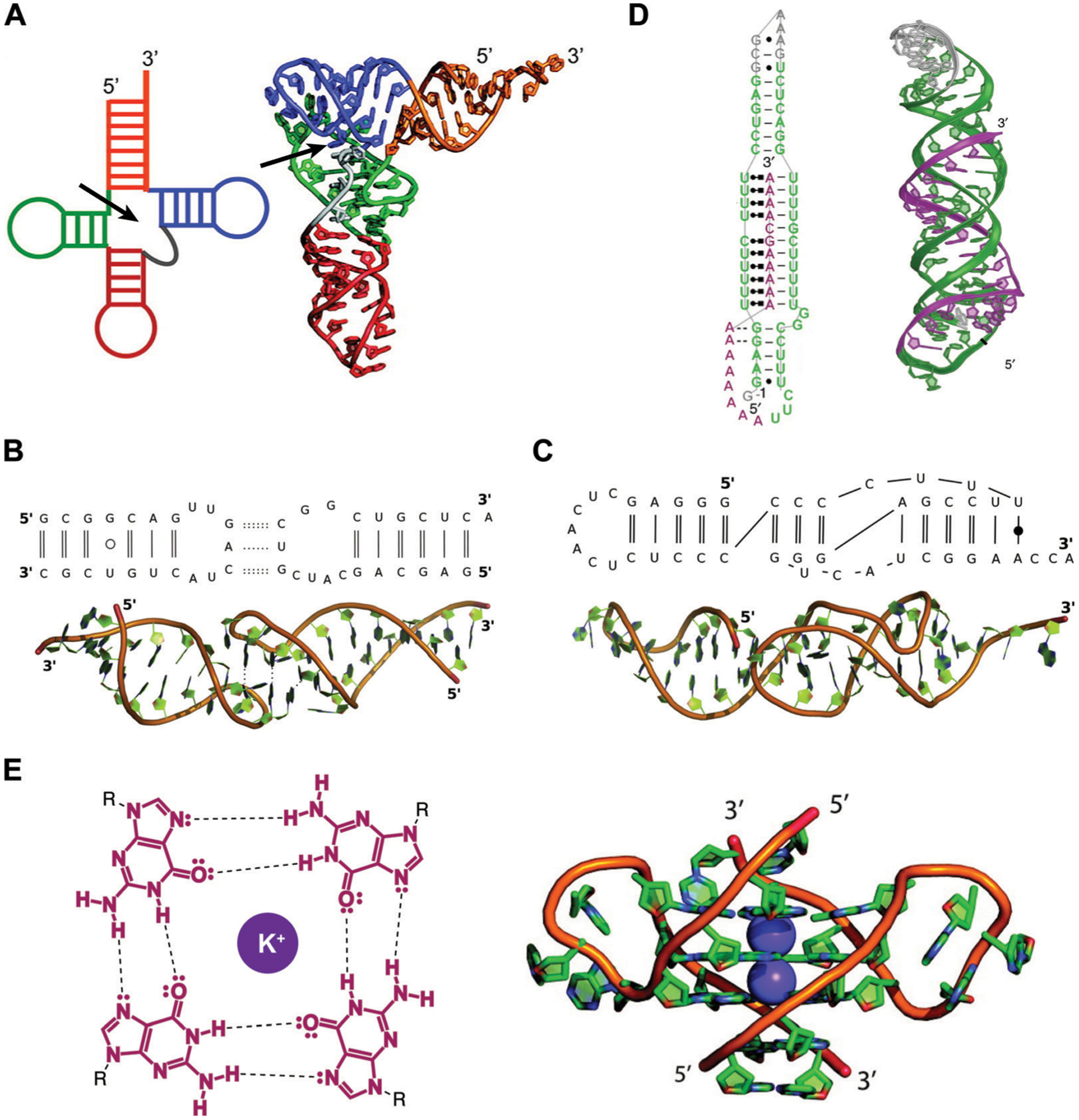
Examples of RNA tertiary structures. (A) Yeast phenylalanine t-RNA (PDB ID: 6TNA). The individual helices arising from the multi-way junction (arrow) that form the cloverleaf-shaped secondary structure (left) were found to stack on each other to determine the L-shaped three-dimensional fold (right). Adapted from ref. 16 with permission from American Chemical Society, Copyright 2011. (B) Example of a kissing loop tertiary interaction formed between substrate and catalytic domain stem-loops of the Neurospora Varkud satellite ribozyme (PDB ID: 2MIO). Dashed lines indicate hydrogen bonding. Wobble and non-WCF base pairs are shown as unfilled and filled circles, respectively. Adapted from ref. 15 with permission from Elsevier B. V., Copyright 2015. (C) Example of a pseudoknot tertiary interaction in the turnip yellow mosaic virus RNA (PDB ID: 1A60). Non-WCF base pair is shown as a filled circle. Adapted from ref. 15 with permission from Elsevier B. V., Copyright 2015. (D) 3′-Triple helix in lncRNA MALAT1 (PDB ID: 4PLX). Uridine-rich stem loop (green) form WCF (simple lines) and Hoogsteen (dot and square lines) base pairs with the A-rich tail (purple). Adapted from ref. 7 with permission from Springer Nature, Copyright 2014. (E) G-quartets are stabilized by potassium cations through interactions with the guanine’s oxygens (left), and stack on top of each other to form G-quadruplex structures (right), as seen in the human telomeric (TERRA) RNA (PDB ID: 3IBK). Dashed lines represent hydrogen bonds, dots represent lone pairs of electrons, and purple circles represent the potassium ion. Adapted from ref. 16 with permission from American Chemical Society, Copyright 2011.
