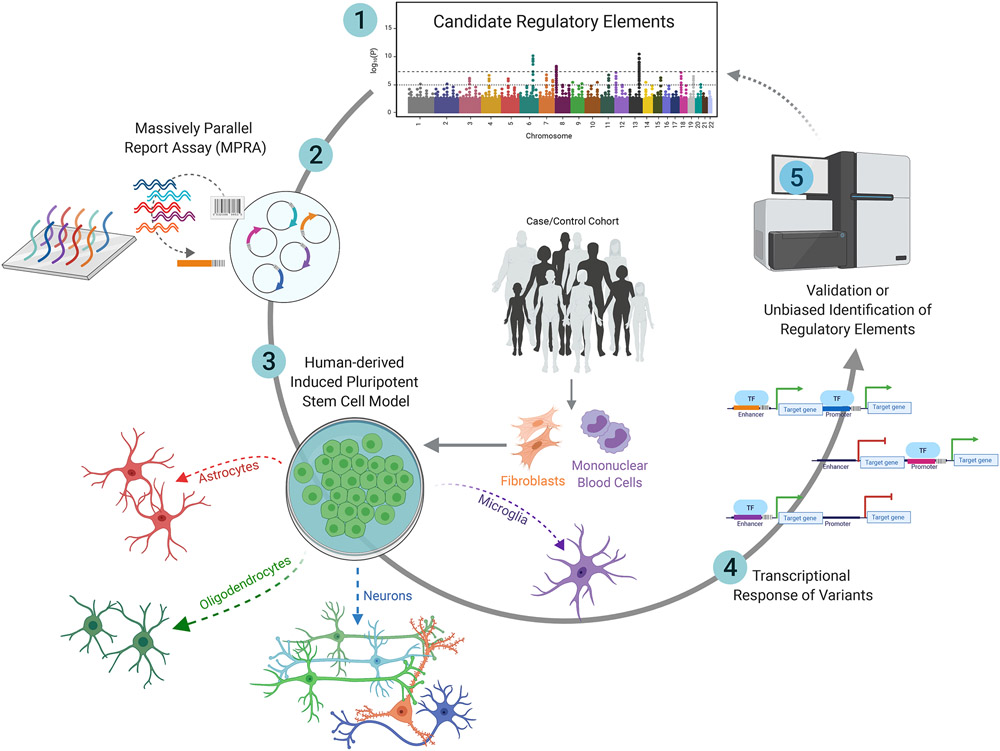
Figure 1: Framework for using massively parallel reporter Assays (MPRAs) to characterize putative regulatory elements with cell-type specificity.
An MPRA library can be designed to include disorder-associated SNPs within putative regulatory elements that have been prioritized through genome-wide association studies (GWAS) and Transcriptomic Imputation (TI) analyses. Patient fibroblasts or peripheral blood mononuclear cells (PBMCs) can be induced into a pluripotent state (hiPSCs). With further adaptation, lenti-MPRA libraries could be integrated into hiPSC-derived mature brain cells. By comparing DNA and RNA sequencing of these cells, MPRAs provide a readout of transcriptional differences between the alleles present in patient versus control populations. If transcriptional shifts exist between variants at a specific region, the transcriptional influence of that SNP can be characterized in the context of genetic risk for a disorder.