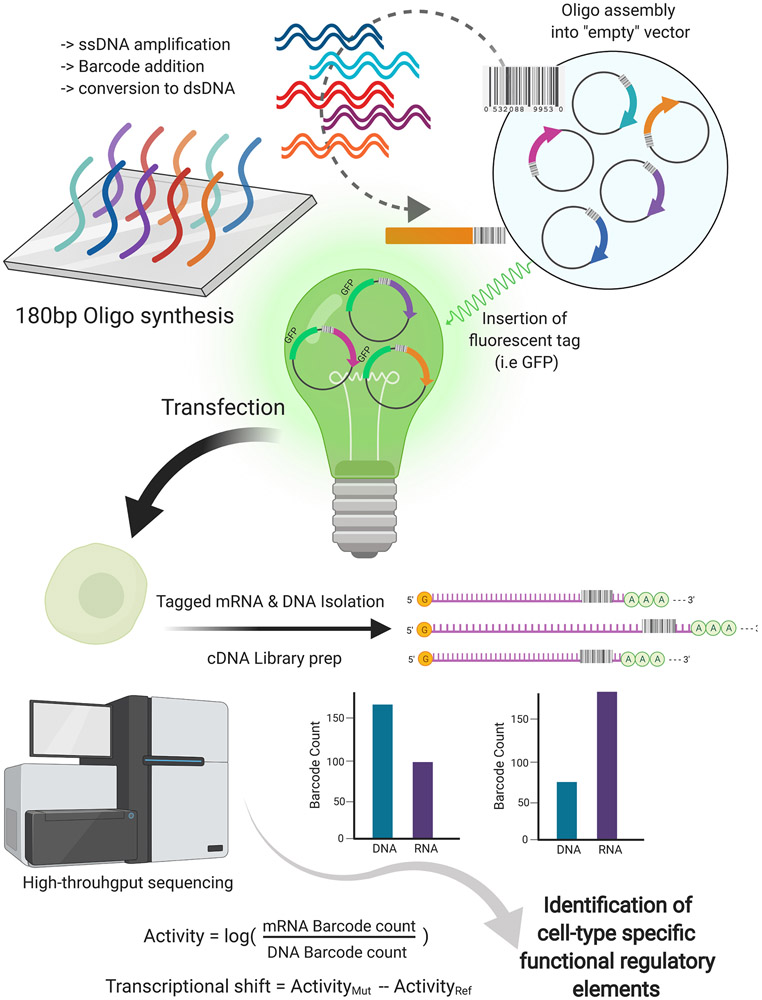
Figure 3: Outline of the Massively Parallel Reporter Assay (MPRA) workflow.
Short 180bp oligos are synthesized by cleavage of single-stranded DNA (ssDNA) from the array. Through emulsion PCR, the 180bp ssDNA oligomers are amplified, barcoded, and converted to double-stranded DNA (dsDNA). dsDNA oligomers are assembled into an empty report vector, creating an MPRA library. The plasmids are fluorescently tagged by the insertion of a GFP open reading frame and minimal promoter for transfection into the desired cell type. RNA is isolated from transfected cells and the barcoded mRNAs are captured and sequenced. Barcode counts are compared to the count estimates from the sequencing of the plasmid library or sequencing of DNA captured simultaneously with RNA to measure relative expression. Sequencing results from MPRAs can be integrated into sequencing information from other techniques, such as ATAC-seq, ChIP-seq, and Capture Hi-C. Application of massively parallel and combinatorial methods in hiPSC models enables the cell-type specific identification of novel regulatory elements as well as the validation of unconfirmed candidates. [ATAC-seq: Assay for Transposase-Accessible Chromatin using sequencing Capture Hi-C: A chromatin conformation capture technique that is target to specific loci; and ChIP-seq: Chromatin immunoprecipitation followed by sequencing]